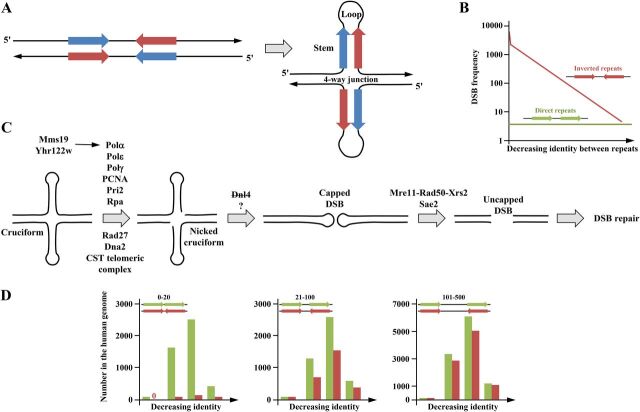
FIG 4.
Inverted repeats and cruciforms. (A) The unstructured palindrome is shown as complementary pink and blue arrows. In certain conditions, it may adopt a cruciform structure containing two stems and two loops of variable lengths that form a four-way junction whose structure is similar to a Holliday junction. The stability of a cruciform depends on both the stem and loop lengths. (B) Double-strand break (DSB) frequency according to the identity between direct or inverted tandem repeats in yeast. More identical repeats are more prone to form stable cruciforms, therefore inducing more frequent DSB. (Based on data from reference 96.) (C) Genetic factors involved in cruciform processing in yeast. The proteins involved in the transition between the nicked form and the capped DSB are not clearly characterized, but ligase IV (DNL4 in yeast) does not play a role in this reaction (96). (D) The number of direct or inverted Alu repeats in the human genome. The distances between repeats were classified into close (0 to 20 nucleotides [nt], left panel), medium (21 to 100 nt, middle panel), and distant (101 to 500 nt, right panel). In each panel, repeats are classified from left to right in order of decreasing identity (>90%, 81 to 90%, 71 to 80%, and 61 to 70% identity). Close inverted repeats are counterselected because they may form stable cruciforms (left panel), whereas this selection is less pronounced at longer distances. (Based on data from https://www.niehs.nih.gov/research/resources/databases/alu/index.cfm.)