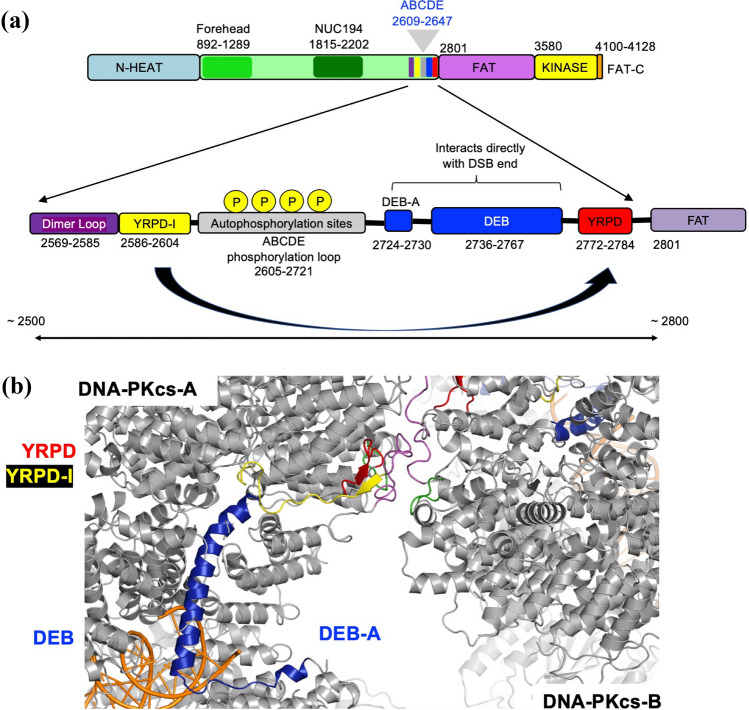
Fig. 4.
Cartoon showing regulatory sequences located in DNA-PKcs between 2500 and 2800. A Schematic showing the region of DNA-PKcs between amino acids 2500 and 2800 that regulates the synaptic interface. The dimerization loop (residues 2569–2585), the YRPD-interacting motif (2586–2604), the DNA End Binding (DEB) helix (2736–2767) with DEB-appended (DEB-A, residues 2724–2730) and a short linker (2731–2735), and the YRPD motif (2722–2784) are from Chen et al. (2021a). DEB-A, the linker and 37 amino acid DEB form a basic surface that interacts directly with the ends of the DSB (Chen et al., 2021a). B Close up view of interaction interface between two DNA-PKcs molecules, labeled DNA-PKcs-A and DNA-PKcs-B, showing the forehead loop (residues 896–903, green), the dimerization loop (2569–2585, purple), the YRPD-I (residues 2586–2604, yellow), the YRPD (2774–2784, red) and the DEB-A, linker DEB (2724–2767, blue). The position of the DEB is maintained through interaction of the YRPD and the YRPD-I, shown as yellow and red beta sheets respectively, the dimerization loop and the forehead loop. The disordered loop containing the ABCDE autophosphorylation sites (residues 2605–2721) is absent from the structures and is not shown