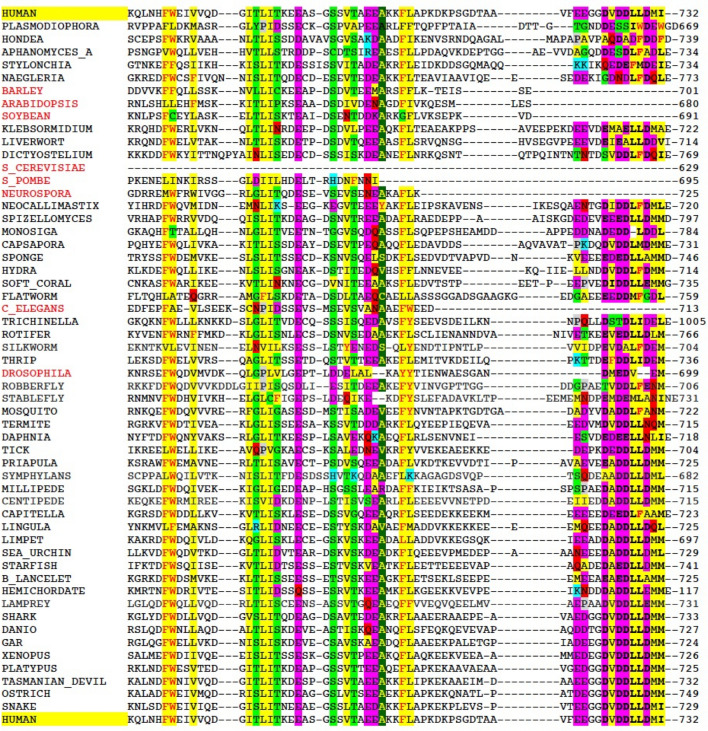
Fig. 6.
Conservation of the C-terminal region of Ku80 in vertebrates and multiple invertebrates and plants that also contain DNA-PKcs, but not in Dikarya yeast, C. elegans and flowering plants that do not. Multiple protein sequence alignment of Ku80 was carried out using Clustal Omega and conserved amino acids were colored manually using the following criteria: acidic amino acids (D, E) are highlighted in magenta; basic amino acids (R, K, H) are highlighted in blue; small polar amino acids (T, S, G, C) are in green; aliphatic amino acids (A, L, M, I, V) are in yellow; large polar amino acids (Q, N) are in red; aromatic amino acids (F, W, Y) are in red with yellow highlight; and proline (P) is in grey. Conservation of small amino acids (A, S, T, C, V, G) is illustrated by yellow letters with dark green highlight. In bold are the amino acids present in the structure of the long-range complex (DVDDLLDM) that interact with DNA-PKcs. Organisms and accession numbers for the sequences shown are as follows: PLASMODIOPHORE (Club root pathogen), Plasmodiophora brassicae, CEO94558.1; HONDAEA (Protist) Hondaea fermentalgiana, BG27299.1; APHANOMYCESa (Crayfish plague mold) Aphanomyces astaci, XP_009833210.1; STYLONYCHIA (Ciliate) Stylonychia lemnae, CDW74323.1; NAEGLERIA (Amoeba/flagellate), Naegleria gruberi strain, XP_002672413.1; BARLEY, Hordeum vulgare, AEO86624.1; ARABIDOPSIS, Arabidopsis thaliana, NP_564520.1; SOYBEAN, Glycine max, XP_003524779.1; GREEN_ALGAE, Klebsormidium nitens, GAQ85406.1; LIVERWORT, Marchantia polymorpha, PTQ30121.1; DICTYOSTELIUM, Dictyostelium purpureum, XP_637846.1; S_CEREVISIAE, Saccharomyces cerevisiae, NP_013824.1; S_POMBE, Schizosaccharomyces pombe, Q9HGM8.1; NEUROSPORA, (Bread mold), Neurospora crassa, AFM68948.1; NEOCALLIMASTIX (Anaerobic gut fungi), Neocallimastix californiae, ORY73184.1; SPIZELLOMYCES (Chytridomycota fungus), Spizellomyces punctatus, KND04299.1; MONOSIGA, (Choanoflagellate), Monosiga brevicollis, XP_001750285.1; CAPSASPORA (Amoeboid protist), Capsaspora kowczarzaki, KJE95979.1; SPONGE, Amphimedon queenslandica, XP_019851059.1; HYDRA, Hydra vulgaris, XP_012555181.1; SOFT_CORAL, Dendronephthya gigantea, XP_028404243.1; FLATWORM, Macrostomum lignano, PAA63650.1; C_ELEGANS, Caenorhabditis elegans, CAA83623; TRICHINELLA, Parasitic roundworm, Trichinella pseudospiralis, KRZ25527.1; ROTIFER, Brachionus plicatilis, RNA39402.1; SILKWORM, Bombyx mori, XP_037875756.1; THRIP, Frankliniella occidentalis, XP_026281764.1; DROSOPHILA, Drosophila melanogaster, NP_609767.2; ROBBERFLY, Eutolmus rufibarbis, SRR5185497; STABLEFLY, Stomoxys calcitrans, XP_013115123.1; MOSQUITO, Aedes aegypti, XP_001657128.2; TERMITE, Cryptotermes secundus, XP_023718673.1; DAPHNIA, Daphnia magna, XP_032789667.1; TICK, Ixodes scapularis, XP_002405506.2; PRIAPULA, Penis worm, Priapulus caudatus, XP_014668438.1; MILLIPEDE, Craspedosoma sp., GERS01021842.1; CENTIPEDE, Himantarium gabrielis, GCIL01016305.1; CAPITELLA, Annelid worm, Capitella teleta, ELU08335.1; LINGULA, Brachiopod, Lingula anatine, XP_013403161.1; LIMPET, Lottia gigantea, XP_009064003.1; SEA URCHIN, Strongylocentrotus purpuratus, XP_788472.3; STARFISH, Acanthaster planci, XP_022103565.1; LANCELET, Branchiostoma belcheri, XP_019618779.1; HEMICHORDATE, Acorn worm, Saccoglossus kowalevskii, XP_006822840.1; LAMPREY, Petromyzon marinus, xp_032818808; SHARK, Prionace glauca, GFYY0116112.1; DANIO, Danio rerio, NP_001017360.2; GAR, Lepisosteus oculatus, XP_015214714.1; XENOPUS, Xenopus laevis, BAA76954.1; PLATYPUS, Ornithorhynchus anatinus, XP_028928834.1; TASMANIAN_DEVIL, Sarcophilus harrisii, XP_031813944.1; OSTRICH, Struthio camelus australis; XP_009666109.1; SNAKE, Pseudonaja textilis, XP_026563791.1. The names of organisms that do not contain DNA-PKcs are shown in red font. All others contain DNA-PKcs. See (Lees-Miller et al., 2020) for details. The human sequence is shown at the top and bottom in yellow highlight for comparison