FIG 2.
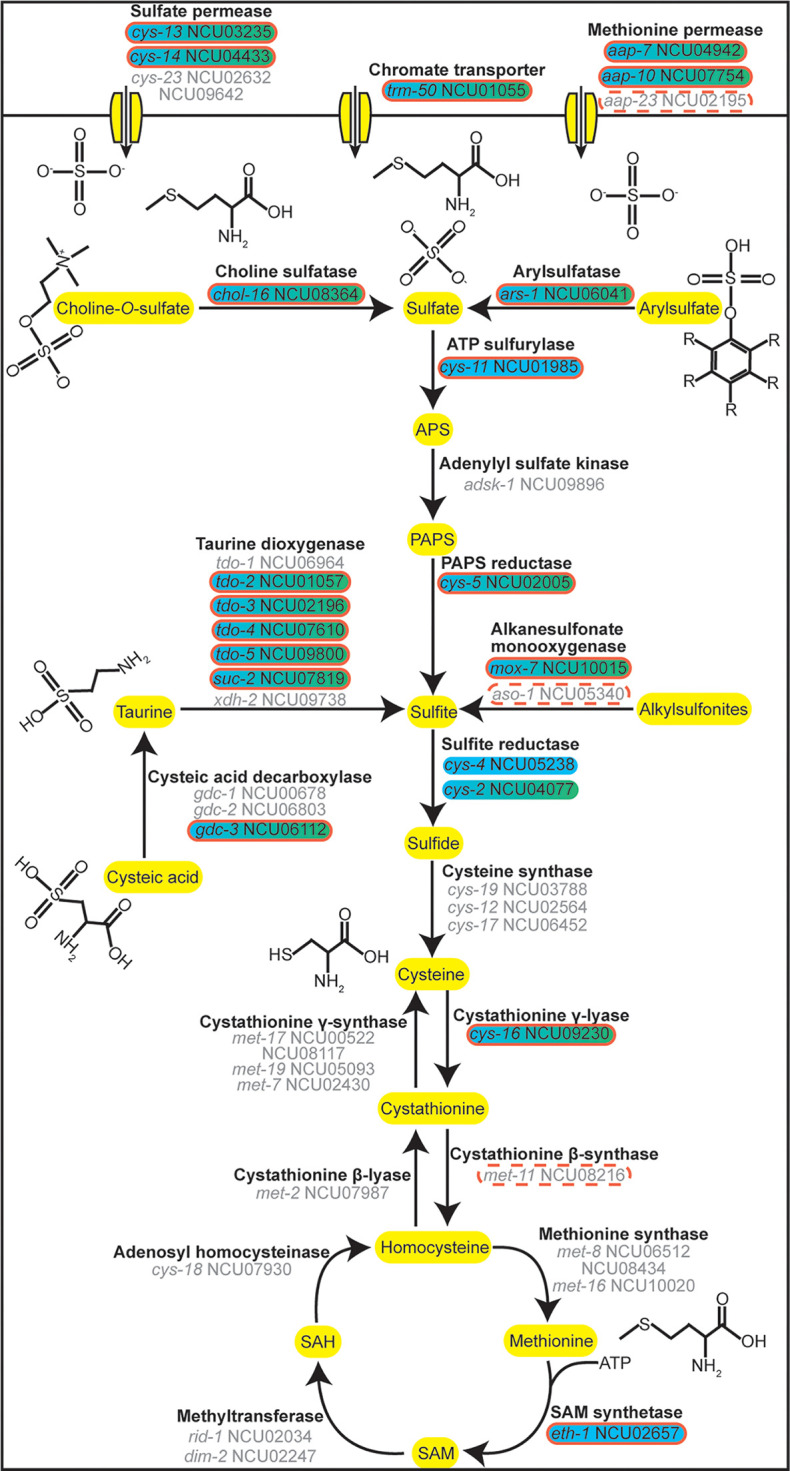
Genes involved in sulfur acquisition but not cysteine and methionine metabolism are regulated by CYS-3 in response to sulfur starvation. Shown is a cartoon of sulfur assimilation in N. crassa. Green shading indicates that the gene was differentially expressed by at least 2-fold in wild-type cells exposed to 24 μM sulfate compared to sulfur starvation. All of these differentially expressed genes were upregulated during sulfur starvation compared to 24 μM sulfate. Blue shading indicates that the gene was differentially expressed by at least 2-fold in wild-type compared to cys-3− cells exposed to sulfur starvation. All of these differentially expressed genes were activated by CYS-3 during exposure to sulfur starvation. Gray text indicates that the gene was not differentially expressed by at least 2-fold under either of the conditions. A solid red outline indicates that CYS-3 bound the gene’s promoter and that the gene was at least 2-fold differentially expressed in wild-type compared to cys-3− cells exposed to sulfur starvation. A dotted red outline indicates that CYS-3 bound the gene’s promoter in the unfiltered DAP-seq data but that the gene was not at least 2-fold differentially expressed under the tested conditions. There was a lower confidence that these promoters were bound by CYS-3 in vivo than promoters of genes with a solid red outline. APS, adenosine 5′-phosphosulfate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine.
