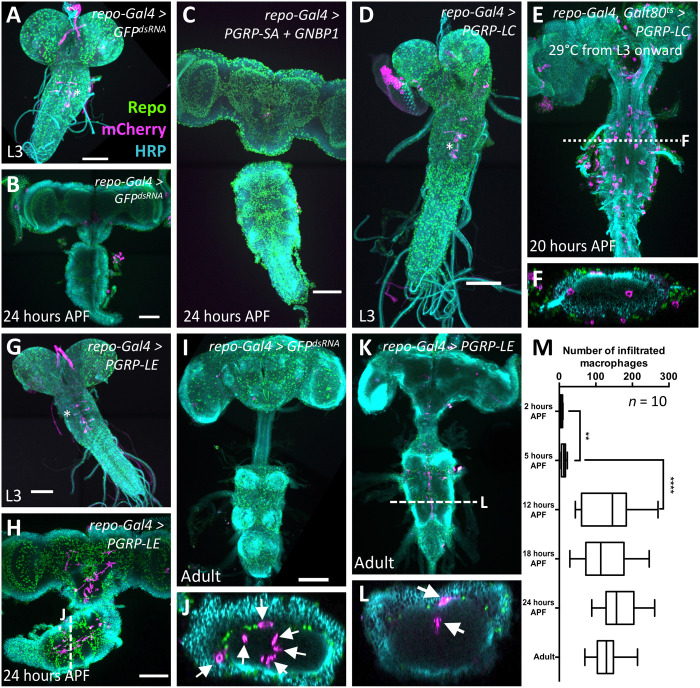
Fig. 2. Macrophages enter the brain during pupal stages upon immunity induction.
Dissected brains of indicated age stained for Repo (green) to label glial nuclei, HRP (cyan) to label neuronal membranes, and mCherry expression (magenta) directed by the macrophage marker srpHemo-moe::3xmCherry. wL3, wandering third instar larva. Adults were 7 days old. Scale bars, 100 μm. (A) Control larva expressing double-stranded RNA directed against green fluorescent protein (GFP). The asterisk indicates six neurons expressing the macrophage marker srpHemo-moe::3xmCherry. (B) Control pupa expressing double-stranded RNA directed against GFP. (C) Pupal brain expressing PGRP-SA and GNBP1 in glial cells. Note the absence of mCherry-expressing cells in the brain. (D) Larval brain expressing PGRP-LC in glial cells, no macrophages are found in the brain. The asterisk denotes neurons weakly expressing the marker srpHemo-moe::3xmCherry. (E) PGRP-LC expression is restricted to late larval and pupal stages. Macrophages invade the brain. The dashed line indicates the orthogonal section shown in (F). (G) Larva with PGRP-LE induction in glial cells. Note the elongated ventral nerve cord. The asterisk indicates neurons weakly expressing the macrophage marker. (H and J) Pupal brain expressing PGRP-LE. Note the presence of srpHemo-moe::3xmCherry–expressing cells in the brain. The white dashed line indicates the position of the orthogonal section shown in (J) (arrows indicate macrophages). (I) Adult control brain expressing GFPdsRNA. (K and L) Adult brain with immunity induction in glia. Cherry-expressing cells are found in the brain. The white dashed line indicates the orthogonal section shown in (L) (arrows indicate macrophages). (M) Quantification of the infiltration rate. To count the number of Cherry-positive cells, we used the srpHemo-H2A::3xmCherry marker. Pupae of increasing age were dissected, and the number of Cherry-expressing cells was determined using Imaris. P values are **P2–5 hours APF = 0.0034 and ****P5–12 hours APF < 0.0001 (t test); n = 10 for every time point.