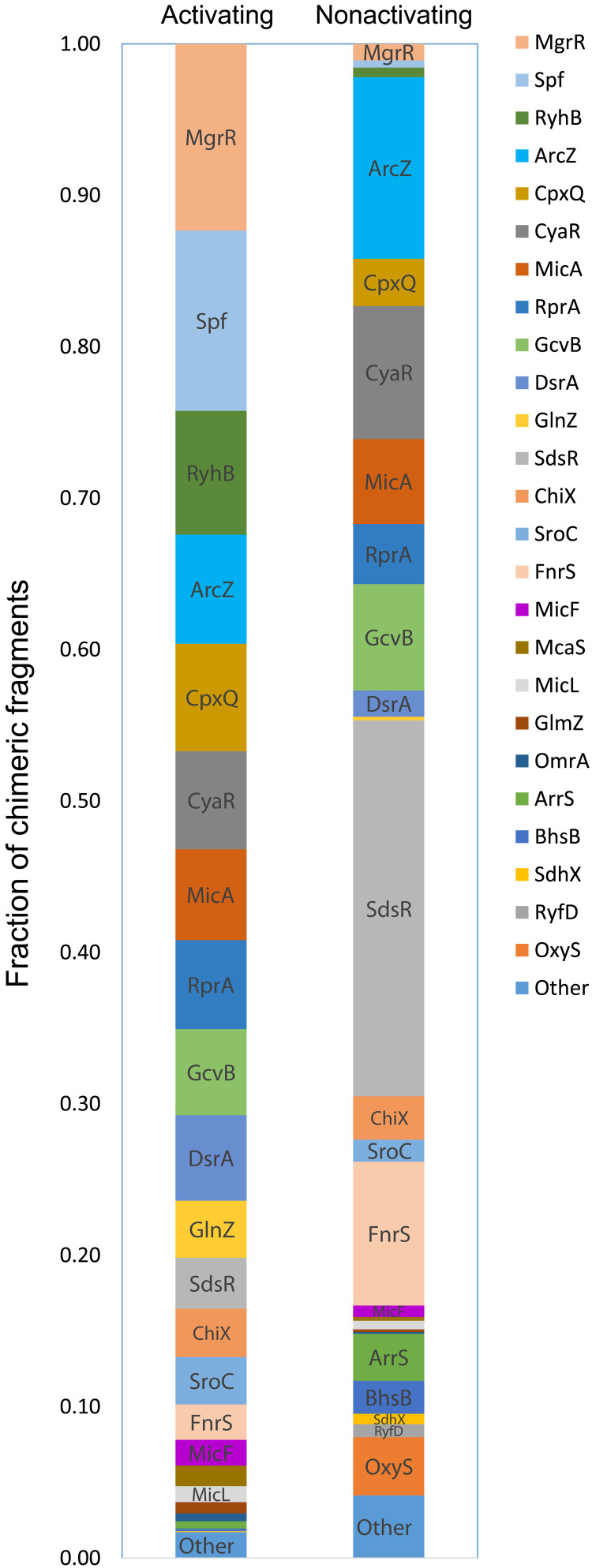
Fig. 2. Relative abundance of core-encoded sRNAs in EPEC RIL-seq data.
For each sRNA in S-chimeras, the relative abundance of the respective chimeric fragments identified in bacteria grown under the activating and nonactivating conditions is shown. This fraction is obtained by dividing the number of chimeric fragments involving a given sRNA by the total number of chimeric fragments involving core-encoded sRNAs. Numbers were extracted from table S3. Some chimeric fragments involve two sRNAs, in which case the number of chimeric fragments is assigned to each of the two interacting sRNAs and the total is updated accordingly. Thus, the total numbers of chimeric fragments used here are 66,540 and 83,865 for activating and nonactivating conditions, respectively. Twenty-six sRNAs with very low abundances under both conditions (<0.005) are grouped to one category termed as “other.”