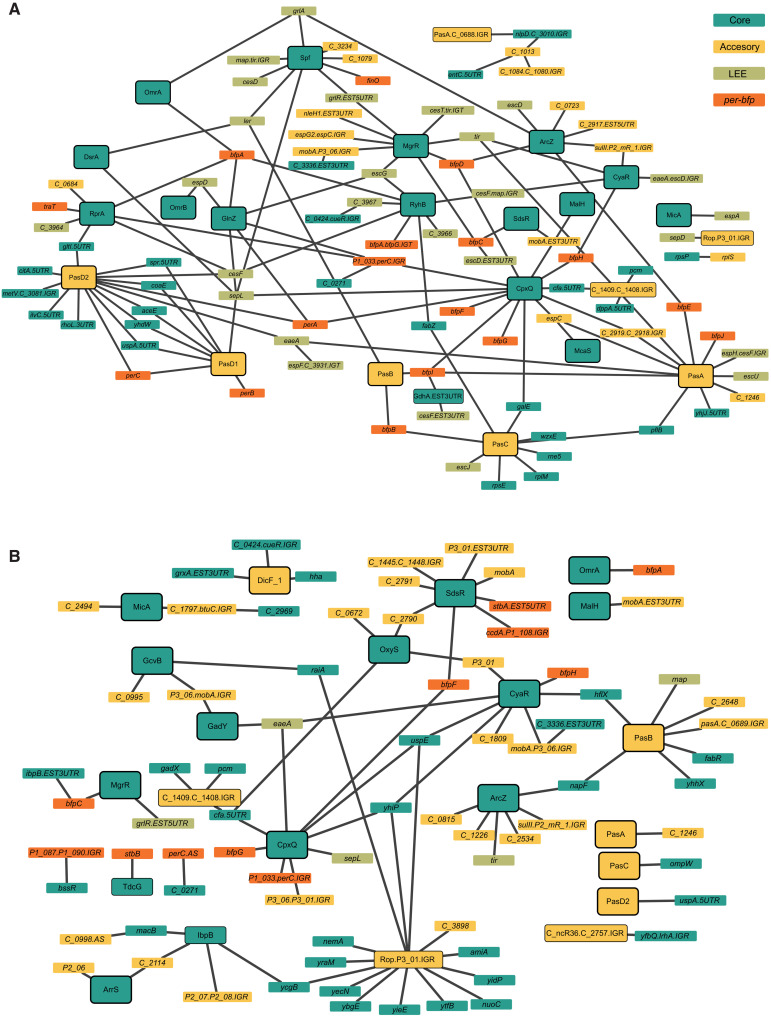
Fig. 5. RIL-seq RNA-RNA virulence-associated interaction networks.
The virulence-associated RNA-RNA interactions identified by RIL-seq under activating (A) and nonactivating (B) conditions are represented as networks, where nodes represent RNAs and edges connect nodes for which an interaction was determined. These networks are filtered for interactions detected in at least two of the triplicate libraries, where for each edge, at least one node is an RNA fragment encoded in the accessory genome. Green, orange, and yellow nodes denote LEE, per-bfp, and other accessory genome transcripts, respectively. Turquoise nodes represent genes encoded in the core genome (Supplementary Material). A node representing a transcript predicted or known to be an sRNA is marked by a black frame.