Figure 3.
Autoinhibition and activation of MINDY1
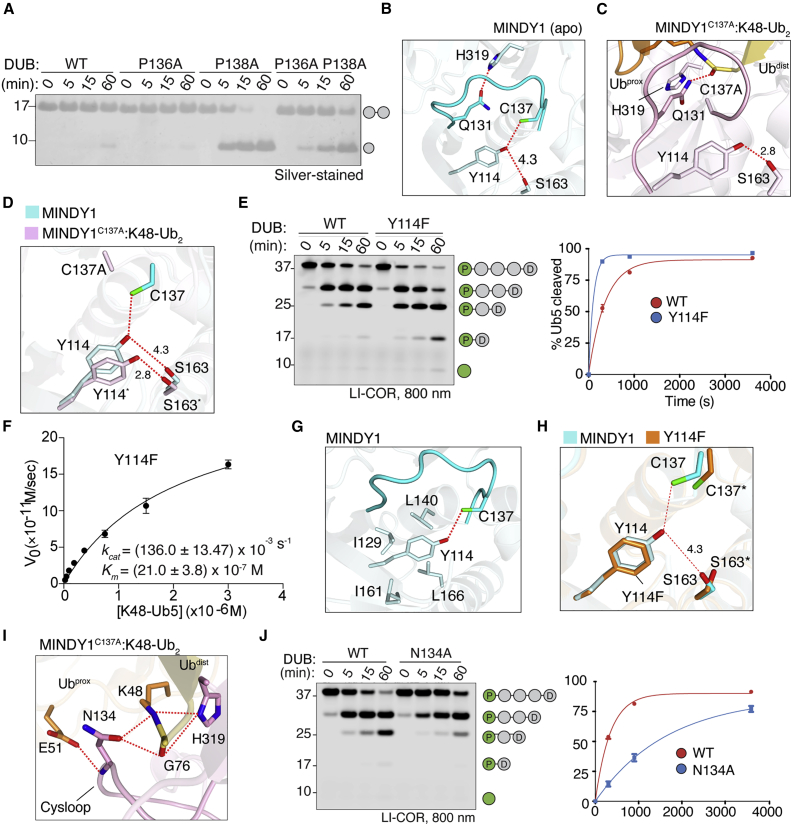
(A) DUB assay monitoring the cleavage of K48-Ub2 by MINDY1 and indicated mutants.
(B) Close-up view of catalytic residues and their interactions in MINDY1 (apo). C137 is out of plane with H139 and is hydrogen bonded with Y114 in MINDY1 (apo). Red dotted lines indicate hydrogen bonds.
(C) Close-up view as in (B) for the MINDY1C137A:K48-Ub2 complex. The catalytically productive state conformation leads to the formation of new sets of bonds as shown. The oxyanion hole residue Q131, which was in contact with catalytic H319 in (B), now forms interactions with the carbonyl of the incoming scissile bond.
(D) Lateral movement of Y114 and its interactions in MINDY1 (apo) and MINDY1C137A:K48-Ub2 complex.
(E) DUB assays comparing cleavage of fluorescently labeled pentaUb by MINDY1 and Y114F mutant. The percentage hydrolysis of pentaUb is plotted against time (right).
(F) Steady-state kinetics of K48-linked pentaUb cleavage by MINDY1 Y114F (n = 2; means ± SDs).
(G) Close-up view of Y114 (phenyl ring) interactions with hydrophobic residues on adjoining secondary structure elements in MINDY1 (apo).
(H) A close-up image of active site of apo MINDY1 Y114F mutant compared to WT. Hydrogen bonding of C137 to Y114 is broken in the mutant.
(I) Interactions of Cys loop residue N134 in stabilizing the isopeptide bond for catalysis.
(J) Cleavage of pentaUb chains fluorescently labeled on Ubprox by MINDY1 WT and N134A mutant. The panel on the right shows the quantification of the DUB assay. n = 2; mean ± SD.
See also Figure S4.