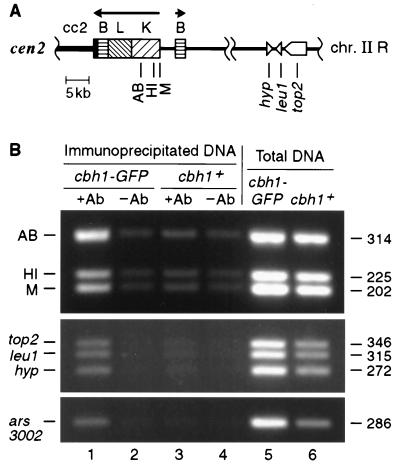
FIG. 4.
GFP-tagged Cbh1p is associated with both centromeric and noncentromeric chromatin in vivo. (A) Schematic of a portion of chromosome II shows the locations that were examined for association with Cbh1p-GFP using a ChIP technique (51, 52). Besides ars3002 and fragment HI of the centromeric K repeat, which were tested as likely cognate targets because they bind Cbh1p in vitro, several other random noncoding and coding sequences were also tested. PCR primer sets are indicated by vertical lines. Fragments AB, HI, and M are located within the centromeric K repeat that is found in 16 to 18 copies in the S. pombe genome. The single-copy genes top2, leu1, and a hypothetical open reading frame (hyp) are located greater than 300 kb from cen2 (Sanger Centre S. pombe Genome Project [http://www.sanger.ac.uk]). The location of ars3002, a telomere-proximal unique sequence on chromosome III, is not shown. Centromeric DNA elements are indicated by solid or patterned boxes. Genes are marked by open, pointed symbols, with the direction of transcription proceeding toward the tip. (B) Coimmunoprecipitation of centromeric K repeat DNA, ars3002 DNA, and random transcribed DNA with Cbh1p-GFP. Cross-linked chromatin from an integrated cbh1-GFP (SBP071198-2) or cbh1+ (SBP040398) strain was immunoprecipitated with antibody (+Ab) against GFP (lanes 1 and 3, respectively) or beads only (−Ab) (lanes 2 and 4, respectively). Total DNA (lanes 5 and 6) and immunoprecipitated DNA were used for PCR with primers specific for the indicated fragments (see Materials and Methods). The corresponding densitometry values for immunoprecipitated DNA normalized to total DNA from cbh1-GFP and cbh1+ strains, respectively, are as follows: AB, 0.707 and 0.192; HI, 0.385 and 0.167; M, 0.391 and 0.147; top2, 0.268 and 0.184; leu1, 0.255 and 0.196; hyp, 0.253 and 0.212; and ars3002, 0.253 and 0.212. The size of each amplified fragment is indicated in base pairs.