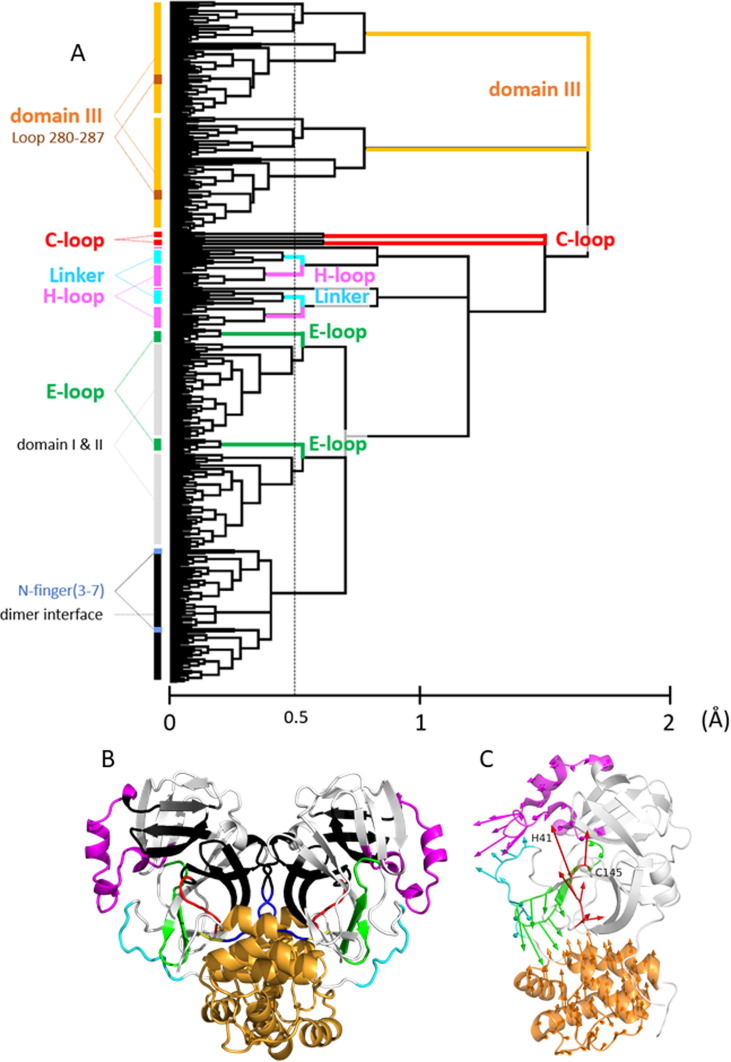
Figure 2.
(A) Motion Tree, a hierarchical clustering of the variance of Cα distances27, 28, calculated for the crystal structure ensemble consisting of all 258 chains of SARS-CoV 3CLpro and SARS-CoV-2 3CLpro. The variances Dmn were symmetrized for two chains, or Dmn = DMN (residues m and n in a protomer correspond to residues M and N in the other protomer, respectively), to treat the homodimer of 3CLpro; thus, two sets of equivalent clusters corresponding to each protomer exist in the tree (see Methods). A taller branch with a high Motion Tree (MT) score, the scale of the abscissa, indicates a cluster moving to a greater extent. The amplitudes of motions follow the order: domain III > C-loop > E-loop ∼ H-loop ∼ Linker. The clusters over the threshold (a thin broken vertical line; MT score = 0.5 Å) are defined as the moving clusters. Nevertheless, domain III is treated as a single cluster for simplicity. Based on the Motion Tree, we defined five moving clusters (the C-loop, E-loop, H-loop, Linker, and domain III). The N-finger (residues 3–7) is shown to be a part of the core of the dimer interface region. Highly mobile Ser1 and Gly2 and the C-terminal residues 301–306 are not included in the analysis because their coordinates are often involved in missing residues. Loop 280–287 belongs to domain III (see discussion in “Influences of T285A mutation …”). (B) The moving clusters of SARS-CoV-2 3CLpro (PDB: 6lu7 as a representative). The color scheme for the moving clusters defined by the Motion Tree (A) is as follows: the C-loop, red; E-loop, green; H-loop, magenta; Linker, cyan; and domain III, bright orange. The core regions consist of the core part of domains I and II (white) and the dimer interface region (black). The C-terminal part of the N-finger (residues 3–7; blue) is within the dimer interface region, whereas residues 1 and 2 (yellow) are highly flexible. Notably, each of the N-termini is located near the C-loop of the other protomer. (C) The first principal components (PCs) of the moving clusters within a protomer are illustrated by arrows using the same color scheme used in (B). The variances explained by PC1 are 0.59 (C-loop), 0.58 (E-loop), 0.35 (H-loop), 0.41 (Linker), and 0.53 (domain III). The catalytic dyad, Cys145 and His41, is depicted by spheres at Cα atoms.