Abstract
Multiplexed detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rather than detection targeting a single gene is crucial to ensure more accurate coronavirus disease 2019 (COVID-19) diagnostics. Here, we develop a monolithic, 3D-printed, lab-on-disc platform for multiplexed molecular detection of SARS-CoV-2. The centrifugal lab-on-disc is fabricated in one step using simple 3D printing technology, circumventing the need for aligning and binding multiple layers. By combining isothermal amplification technology, this lab-on-disc platform is capable of simultaneously detecting the nucleoprotein and envelope genes of SARS-CoV-2 as well as an internal control of the human POP7 gene. Within a 50-minute incubation period, 100 copies SARS-CoV-2 RNA can be detected through visual observation according to color and fluorescence changes in the disc. Further, we clinically validated the lab-on-disc platform by testing 20 nasopharyngeal swab samples and demonstrated a sensitivity of 100% and an accuracy of 95%. Therefore, the monolithic, 3D-printed, lab-on-disc platform provides simple, rapid, disposable, sensitive, reliable, and multiplexed molecular detection of SARS-CoV-2, holding promise for COVID-19 diagnostics at the point of care.
Keywords: COVID-19, SARS-CoV-2, Multiplexed molecular detection, Lab-on-disc, 3D printing
Graphical Abstract
1. Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has claimed more than 4 million lives all over the world [1]. Although an increasing number of COVID-19 vaccine doses are being administered in many countries, rapid SARS-CoV-2 diagnostics of presymptomatic and asymptomatic COVID-19 patients still plays a vital role in combating the COVID-19 pandemic [2], [3].
Currently, the most commonly used SARS-CoV-2 diagnostics approach is reverse transcription polymerase chain reaction (RT-PCR) [4], [5]. By targeting a single gene or multiple genes, RT-PCR can identify SARS-CoV-2 with high sensitivity and specificity [6], [7]. However, RT-PCR assays depend on expensive thermal cycling instruments, require over 2 h to complete, and must be conducted by well-trained personnel in central laboratories, and thus are often inaccessible in rural areas and resource-limited settings. As alternatives to RT-PCR assays, reverse transcription isothermal amplification assays such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) [8], reverse transcription recombinase polymerase amplification (RT-RPA) [9], and reverse transcription dual-priming mediated isothermal amplification (RT-DAMP) [10] offer advantages of rapid testing, ease of use, low cost, independence of bulky instruments, and visual product detection. To improve the detection accuracy, researchers have begun developing isothermal amplification assays targeting multiple genes of SARS-CoV-2 [11], [12], [13]. However, most of them rely on multiple fluorescence probes and bulky expensive fluorescence detector, which is not ideal for point of care diagnostic application in resource-limited settings. Therefore, rapid, low-cost, disposable, and multiplexed molecular detection platform of SARS-CoV-2 remains a highly unmet need.
To this end, we sought to leverage a lab-on-disc platform, which make it possible to couple centrifugal microfluidic discs and isothermal amplification. Towards point-of-care testing, lab-on-disc platforms have particular advantages [14], [15]. First, a low-cost spinning motor drives the fluid flow in the discs, avoiding the need for complicated pneumatic interfaces and pumps. Second, centrifugal forces can efficiently remove disturbing bubbles and residual volumes of samples and reagents. Third, multiple chambers can be fabricated to achieve multiplexed assays easily. Current fabrication methods for lab-on-discs typically involve aligning and binding processes for multiple layers, such as soft lithography, hot embossing and injection molding, and micromachining [16], [17], [18]. However, such time-consuming and bulky instrumentation-dependent fabrication approaches challenge the molecular diagnostic applications of lab-on-disc platforms. The emerging 3D printing technology provides an alternative method to rapidly manufacture complex microfluidic devices for bioassays [19], [20]. With the advancement of inexpensive benchtop 3D printers with high-resolution (down to 25 µm) capabilities, researchers have successfully applied 3D printed microfluidic devices to pathogen detection, blood separation, electrochemical sensors, cell culture, and more.
In this study, we developed a monolithic, 3D-printed, and inexpensive lab-on-disc platform for rapid, multiplexed, molecular detection of SARS-CoV-2. The lab-on-disc platform can simultaneously detect three different genes including the nucleoprotein (N) and envelope (E) genes of SARS-CoV-2 as well as an internal control of the human POP7 gene (POP7). The detection results are visually inspected based on color and fluorescence changes in the discs. Further, we have clinically validated the lab-on-disc platform by testing clinical nasopharyngeal (NP) swab samples. Thus, the 3D-printed lab-on-disc platform provides simple, rapid, disposable, sensitive, reliable, and multiplexed SARS-CoV-2 detection, offering great potential to facilitate COVID-19 diagnostics at the point of care.
2. Experimental section
2.1. Chemicals and materials
Betaine (5.0 M), calcein (5 G), manganese (II) chloride tetrahydrate (100 G), and dimethyl sulfoxide (DMSO) were purchased from MilliporeSigma (Burlington, MA). EvaGreen dye (20 ×) was purchased from Biotium (Fremont, CA). Microseal 'B' PCR Plate Sealing Film, adhesive, optical #MSB1001, and mineral oil #1632129 were purchased from Bio-Rad Laboratories (Hercules, CA). The QIAamp Viral RNA Mini Kit (50) was purchased from Qiagen (Hilden, Germany). Nuclease-free water, dNTP mix (10 mM of each), 10 × isothermal amplification buffer, MgSO4 (100 mM), extreme thermostable single-stranded DNA binding protein (ET SSB; 500 µg mL-1), Bst 2.0 WarmStart DNA polymerase (Bst 2.0 WS; 8000 U mL-1), and WarmStart RTx reverse transcriptase (WS RTx; 15,000 U mL-1) were purchased from New England BioLabs (Ipswich, MA). The GoTaq Probe 1-Step RT-qPCR kit was purchased from Promega (Madison, WI). The synthetic SARS-CoV-2 RNA control was purchased from Twist Bioscience (South San Francisco, CA). POP7 plasmid, SARS-CoV N plasmid control, MERS-CoV N plasmid control, and all of the primers were purchased from Integrated DNA Technologies (Coralville, IA). A total of 20 de-identified, clinical NP swab samples were handled in compliance with ethical regulations and the approval of the Institutional Review Board of the University of Connecticut Health Center (protocol #P61067).
2.2. 3D printing of the lab-on-disc platform
A high-resolution stereolithographic (SLA) laser-based 3D printer Form 2 from Formlabs was used to print the disc. The material for 3D printing was a clear methacrylate-based resin (Formlabs, FLGPCL02). First, a 3D design drawing of the disc was created using SolidWorks software (Waltham, MA). After uploading the drawing file, the printer began to work at a 50 µm resolution. Upon completion, the printed discs were removed from the build plate in the printer and immersed in isopropanol for 20 min, followed by a 25-min ultrasonic cleaning with acetone. The printed centrifugal lab-on-disc was then thoroughly washed with deionized water and rapidly dried with air. All of the lab-on-discs were stored in a cool, dry place until use.
2.3. Tube-based visual RT-DAMP assays
A calcein-MnCl2 dye was used to develop the visual RT-DAMP assays. A 500 mM MnCl2 solution was prepared in DMSO and diluted to 50 mM using DMSO. Similarly, a 2.5 mM calcein solution was prepared with DMSO. After mixing, a calcein (1.25 mM)-MnCl2 (25 mM) dye was prepared. A typical 25 μL visual RT-DAMP assay included 1 × Isothermal Amplification Buffer, 0.2 μM forward outer primer (FO), 0.2 μM reverse outer primer (RO), 1.6 μM forward inner primer (FI), 1.6 μM reverse inner primer (RI), 1.6 μM forward pairing-competition primer (FC), 1.6 μM reverse pairing-competition primer (RC), 0.3 U/μL WS RTx, 1.2 U/μL Bst 2.0 WS, 0.5 μL of the calcein (1.25 mM)-MnCl2 (25 mM) dye, 2.5 ng/μL ET SSB, 0.8 M betaine, and 1.0 μL target solution. The reactions were incubated at 63 °C for 50 min in a Bio-Rad CFX96 Touch real-time PCR detection system. All primer sequences are shown in Table S1. RT-DAMP primers were designed according to principles previously reported by our lab [21]. After incubation, the tubes were placed on a black background or in a Bio-Rad gel imaging system with ultraviolet (UV) light for endpoint visual detection.
2.4. Operation and assays of the lab-on-disc platform
Before operating, adhesive sealing films (Microseal 'B' Film) were clipped to proper sizes to fully cover the lab-on-disc surfaces. Each primer set was pipetted into every two adjacent chambers. For each chamber, 2.5 μL of 10 × primer mix was added, followed by 7.5 μL nuclease-free water. The 10 × primer mix consisted of 2 μL of 100 μM FO, 2 μL of 100 μM RO, 16 μL of 100 μM FI, 16 μL of 100 μM RI, 16 μL of 100 μM FC, 16 μL of 100 μM RC, and 32 μL nuclease-free water. After sealing tightly with adhesive film, a short centrifugation (about 5 s) was run to fully load the primers. Then, the sample inlet was cut open and 150 μL of visual RT-DAMP master mix with targets was added. The master mix contained 25 μL of 10 × Isothermal Amplification Buffer, 15 μL of 100 mM MgSO4, 35 μL of 10 mM dNTPs, 2.5 μL of 120 U/μL Bst 2.0 WS, 5 μL of 15 U/μL WS RTx, 1.25 μL of 0.5 μg/μL ET SSB, 40 μL of 5 M betaine, 5 μL of the calcein (1.25 mM)-MnCl2 (25 mM) dye, 10 μL target solution, 10 μL POP7 plasmid solution (optional), and nuclease-free water to reach 150 μL. A piece of adhesive sealing film was used to seal the sample inlet and the disc was subjected to a 5-second centrifugation to load the master mix into each chamber. Subsequently, the sample inlet was teared open and 200 μL mineral oil was carefully added. The oil was used to entirely isolate each chamber driven by centrifugal forces. Finally, the reagent-filled disc was placed on a mini heat block for a 50-min incubation at 63 °C. Visual detection at the endpoint was carried out by using a black background or the Bio-Rad gel imaging system with UV light. The SARS-CoV-2 detection assays were assessed for specificity, sensitivity, and clinical validation. For the specificity assays, SARS-CoV-2 RNA, SARS-CoV N plasmid, MERS-CoV N plasmid, and human RNA extract were used as the target solutions. For the sensitivity assays, the targets were 10-fold serial dilutions of the commercial SARS-CoV-2 RNA (Twist Bioscience), each mixed with 105 copies of POP7 plasmid. Clinical validation was conducted by using 20 de-identified clinical NP swab samples (10 SARS-CoV-2 positives and 10 SARS-CoV-2 negatives).
2.5. Quantitative RT-PCR (RT-qPCR) assays
Real-time fluorescence RT-qPCR assays targeting the SARS-CoV-2 N gene region were performed strictly according to the Centers for Disease Control and Prevention (CDC)-released instructions titled CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel [22]. The CDC-recommended GoTaq Probe 1-Step RT-qPCR kit was employed to prepare the reaction mix. A typical 15 μL RT-qPCR mix included 1 × GoTaq Probe Master Mix, 0.5 μM nCOV_N1 forward primer, 0.5 μM nCOV_N1 reverse primer, 0.125 μM nCOV_N1 probe, 0.3 μL of the GoScript Reverse Transcriptase Mix, and 1.0 μL of the target solution. The thermal cycling had four stages: Stage 1 (2.0 min at 25 °C); Stage 2 (15.0 min at 50 °C); Stage 3 (2.0 min at 95 °C); Stage 4 (45 cycles of 3.0 s at 95 °C and 30 s at 55 °C). The fluorescence capture point was set at 55 °C in Stage 4. Real-time fluorescence detection was implemented in a Bio-Rad CFX96 Touch real-time PCR detection system.
3. Results and discussion
3.1. Overview of the monolithic, 3D-printed lab-on-disc platform
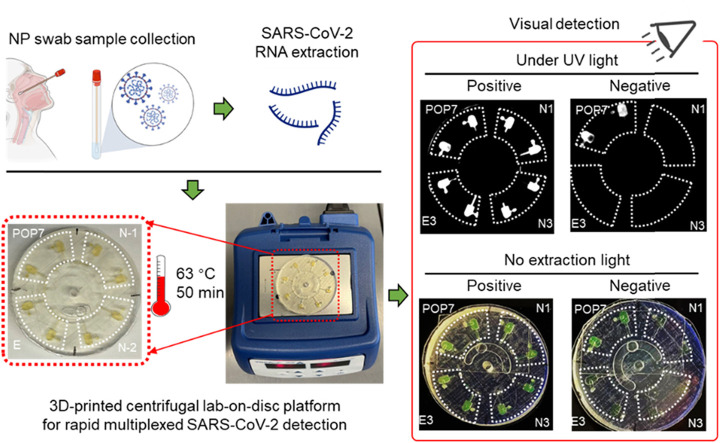
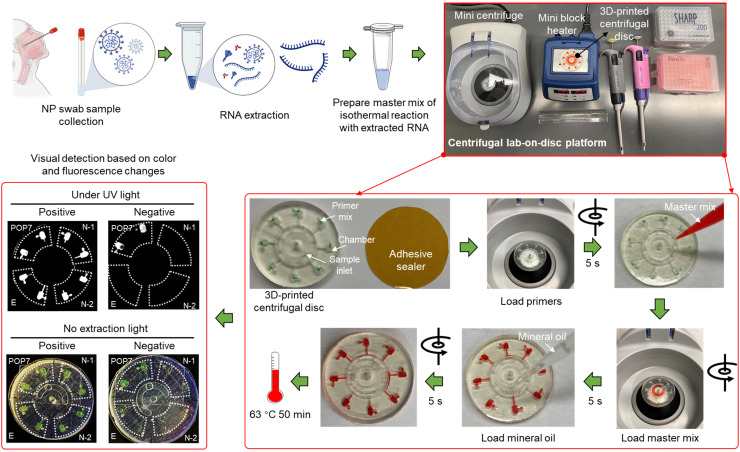
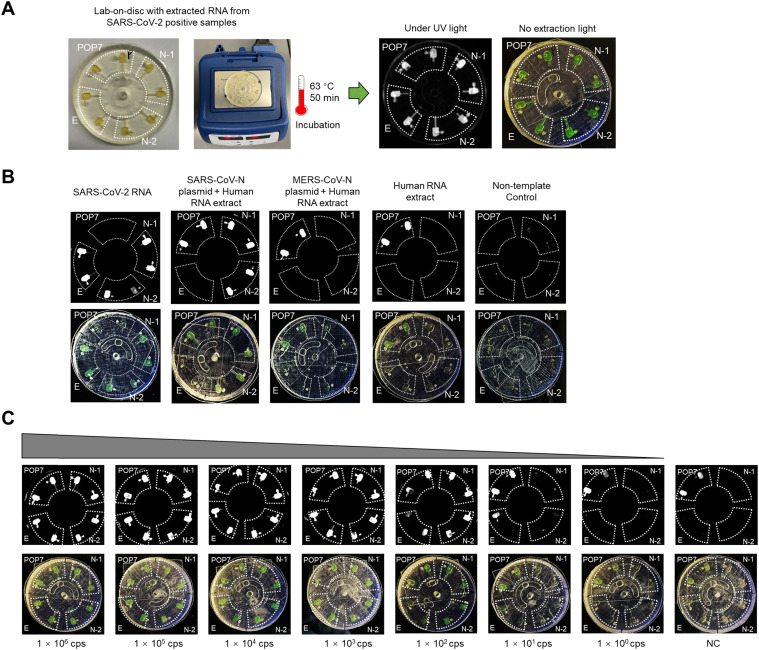
As shown in Fig. 1, the lab-on-disc platform for rapid, multiplexed detection of SARS-CoV-2 mainly consists of the 3D-printed lab-on-disc, a portable mini centrifuge, and a mini block heater. For implementation, clinical NP swab samples are collected from patients, followed by RNA extraction. A master mix of isothermal amplification reaction is prepared by adding the extracted RNA, and is then applied for multiplexed molecular detection of SARS-CoV-2 via the lab-on-a-disc platform. Prior to sealing with adhesive tape, the primer mixes are loaded into the eight chambers. If every two adjacent chambers are filled with the same primer set, the lab-on-disc platform is capable of simultaneously detecting four different target genes. In this study, we have applied the platform to detect two N gene regions (N-1 and N-2) and one E gene region (E) of SARS-CoV-2, as well as an internal control of the human gene region POP7. After a short centrifugation (about 5 s), the primer mixes are entirely loaded into the chambers. Subsequently, the prepared master mix containing the extracted RNA is pipetted into the sample inlet, which is then sealed using a piece of adhesive tape. Similarly, the master mix is distributed to each chamber in a 5-second centrifugation and mineral oil is then loaded for chamber isolation. The sample inlet is sealed again and the disc is incubated at 63 °C for 50 min on the mini block heater. The positive results are visually judged based on the chambers’ color changes from light orange to dark green without excitation light, or the enhanced fluorescence under UV light. Not including the time for RNA extraction, the whole procedure takes only 51 min to obtain detection results. Accordingly, this lab-on-disc platform, independent of bulky instruments, is suitable for rapid, point-of-care testing of SARS-CoV-2 in resource-limited settings.
Fig. 1.
Procedures of the monolithic, 3D-printed lab-on-disc platform for rapid, multiplexed detection of SARS-CoV-2. The NP swab samples are collected first, followed by RNA extraction and master mix preparation. Through employing a mini centrifuge (myFuge™ Mini Centrifuge) and a mini block heater, a centrifugal lab-on-disc platform is formed. To use the platform, the primer mix for each target is first injected into each chamber, then loading the master mix and mineral oil after a very fast centrifuging (about 5 s at 6000 rpm for each). Post 50-min incubation at 63 °C, the detection results can be visually judged under UV light or without extraction light (natural light). NP, nasopharyngeal. N-1, N-2, E, and POP7 are the corresponding primer sets targeting two SARS-CoV-2 N gene regions, one SARS-CoV-2 E gene region, and one human gene region, respectively. Detailed sequence information is shown in Table S1.
3.2. Design and characterization of the monolithic, 3D-printed lab-on-disc platform
Current centrifugal lab-on-disc platforms are most often fabricated using two strategies, the photolithography method and the non-photolithography method [17]. In the former, polymer materials such as polydimethylsiloxane (PDMS) are used to fabricate the lab-on-discs using photolithography-created molds. In the latter, the lab-on-disc is made through the assembly of multiple layers of polymethyl methacrylate (PMMA) and adhesive polyethylene terephthalate (PET) or PDMS. Although both of these approaches can create discs with multifunction integration, the required steps of aligning and binding multiple layers as well as surface modifications greatly complicate the fabrication [16], [23], [24]. Thus, in this study, we employed a simple SLA-based 3D-printing technology for rapid, one-step, prototyping of lab-on-disc systems with high resolution, optical transparency, and minimized roughness.
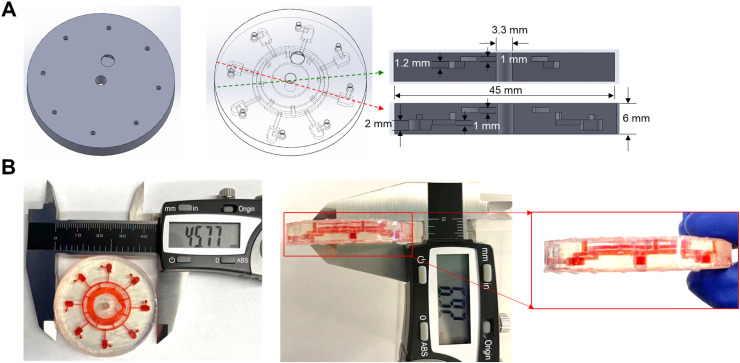
As shown in Fig. 2A, a total of eight chambers (approximately 25 μL for each) at the bottom of the disc are vertically connected with a small circular channel (1.2 mm height) in the middle and a large circular chamber (approximately 200 μL) at the top serves as the reaction mix inlet. In the disc’s center, there is a hollow hole with a diameter of about 3.3 mm, compatible with the spin axis of the mini centrifuge. The printed disc is 45.77 mm in diameter and 6.92 mm in thickness (Fig. 2B). After filling with red dyes, vertical 3D structures are clearly evident in the disc. The lab-on-disc is fabricated in one step by a 3D printer, avoiding the aligning and binding processes of multiple layers. This rapid prototyping procedure is beneficial for large-scale and low-cost production of lab-on-disc systems, enabling fast deployment for scaled COVID-19 diagnostics.
Fig. 2.
(A) Schematic illustration of the monolithic, 3D-printed lab-on-disc. (B) Photograph and size of the 3D-printed lab-on-disc. Red dye was utilized to denote the inner structures of the lab-on-disc. The scale is micrometer (mm).
3.3. Tube-based visual isothermal amplification for SARS-CoV-2 detection
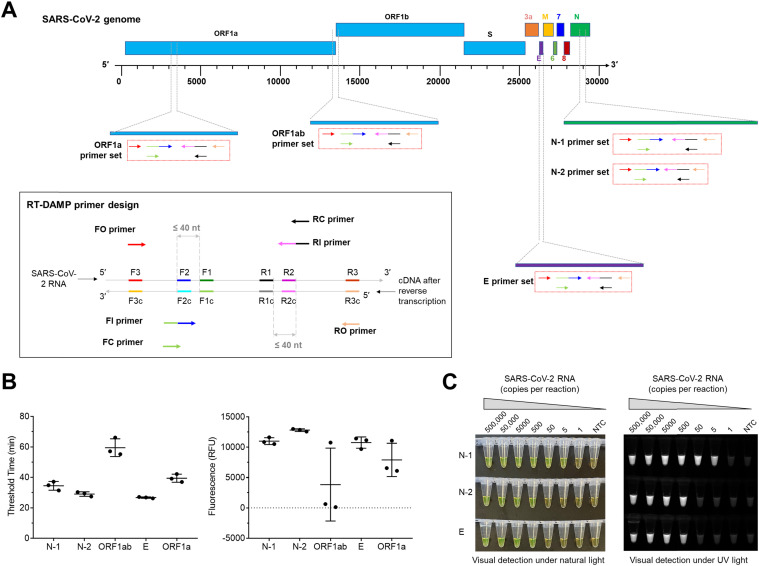
Prior to testing in the lab-on-disc platform, we developed a tube-based visual isothermal amplification approach for SARS-CoV-2 detection by targeting different genes. As shown in Fig. 3A, we investigated a total of five RT-DAMP primer sets targeting N-1, N-2, E, ORF1a, and ORF1b of SARS-CoV-2. The RT-DAMP assay is a variation of RT-LAMP with improved performance developed in our lab. In one-pot RT-DAMP, six primers of FO, RO, FI, RI, FC, and RC (Fig. 3A) specifically recognize six distinct target sites to mediate self-priming and pair-priming (dual-priming) amplification using reverse transcriptase and DNA polymerase. In light of the short threshold time and high endpoint fluorescence (Fig. 3B and S1), we selected the primer sets of N-1, N-2, and E for the subsequent development of our lab-on-disc system. By testing various copies of SARS-CoV-2 RNA, we demonstrated that visual RT-DAMP assays with N-1, N-2, and E primer sets can detect 5, 500, and 500 copies RNA within 50 min, respectively, showing high detection sensitivities (Fig. 3C). Further, we designed an additional primer set targeting the human POP7 gene as an internal control for SARS-CoV-2 detection for the RT-DAMP assay. As shown in Fig. S2, the assay with the internal control is also highly sensitive, as it can detect 500 copies of the POP7 gene sequence. Accordingly, we employed the N-1, N-2, E, and POP7 primer sets to establish the lab-on-disc assay.
Fig. 3.
Tube-based RT-DAMP assays for SARS-CoV-2 visual detection. (A) Primer design of RT-DAMP and genomic locations of primer sets targeting various genes. (B) Comparison of RT-DAMP assays with various primer sets on threshold time in EvaGreen-based real-time detection and the endpoint fluorescence at 50 min. In the assays, 50,000 copies of SARS-CoV-2 RNA were investigated. (C) Sensitivities of visual RT-DAMP detections with N-1, N-2, and E primer sets under natural and UV light. Experiments were repeated for three times. Calcein-MnCl2 dye was used to achieve visual detection and the incubation time was 50 min. NTC, non-template control reaction. Error bars represent the means ± standard deviations from three replicates.
3.4. Lab-on-disc platform for multiplexed, molecular detection of SARS-CoV-2
To combat the ongoing COVID-19 pandemic, a rapid, low-cost, disposable, and multiplexed system for SARS-CoV-2 detection is needed. Previously reported strategies primarily involve multiple fluorescence probes and large detection instruments, which are not appropriate for point-of-care testing [6], [7], [11], [12], [13]. Alternatively, microfluidic chip-based multiple molecular detection systems are more advantageous, due to simplicity and low cost [25], [26]. Here, we developed a monolithic, 3D-printed lab-on- disc platform for rapid, multiplexed molecular detection of SARS-CoV-2. In this platform, the microfluidic discs are fabricated in one step and the centrifugal forces enable fast reagent loading without need for time-consuming pipetting operation.
As shown in Fig. 4A, we pre-filled every two adjacent chambers with one primer set in the disc, thereby simultaneously detecting four different targets (N-1, N-2, E, and POP7). After loading the RT-DAMP master mix with extracted RNA from SARS-CoV-2-positive samples, we incubated the disc at 63 °C for 50 min on a mini heat block. Under UV and natural light, positive chambers indicate strong fluorescence and green color, providing visual multiplexed SARS-CoV-2 detection. Due to the vertical structural design plus light oil sealing, each chamber in the disc is completely isolated without causing reagent coalescence (Fig. S3). Further, we demonstrated that the lab-on-disc platform is specific to SARS-CoV-2 RNA and has no cross-reactivity towards the N gene or human RNA extract of Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) [27] (Fig. 4B). However, the platform has a cross-reactivity with the N gene of SARS-CoV, another fatal coronavirus from 2003 [28]. Through aligning the N gene sequences of MERS-CoV, SARS-CoV, SARS-CoV-2, and four other common human coronaviruses (hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E [29]), we found that SARS-CoV-2 has high genetic homology with SARS-CoV with only few single nucleotide polymorphisms (SNPs) in the corresponding primer sites for the N-1 and N-2 primer sets (Fig. S4 and S5). Similar to RT-LAMP, using RT-DAMP with nonspecific dyes such as EvaGreen, calcein, and pH indicators is difficult to differentiate targets with few SNPs, unless coupling with mismatch-suppression enzymes or specially designed probes [30], [31], [32], [33]. Thus, the specificity of the current lab-on-disc platform would be further improved by using SNP-specific probes. However, although SARS-CoV is also a fatal virus [34], there have not been any known SARS-CoV cases reported globally since 2004. By testing various copies of a commercial SARS-CoV-2 RNA control, we investigated the lab-on-disc platform’s sensitivity. As shown in Fig. 4 C, within 50 min, the platform is able to detect 100 copies of SARS-CoV-2 RNA, showing high sensitivity. Thus, the lab-on-disc platform is specific and highly sensitive at detecting SARS-CoV-2.
Fig. 4.
Lab-on-disc platform for multiplexed molecular detection of SARS-CoV-2. (A) Procedure of the detection platform. (B) Specificity of the detection platform. SARS-CoV-2 RNA, a commercial SARS-CoV-2 RNA control from Twist Bioscience. SARS-CoV-N plasmid and MERS-CoV-N plasmid, commercial products from Integrated DNA Technologies (IDT). Human RNA extract, extracted RNA from a SARS-CoV-2-negative NP sample. (C) Sensitivity of the detection platform using serially diluted SARS-CoV-2 RNA mixed with 105 copies (cps) of POP7 plasmid. The incubation time was 50 min.
3.5. Validation of the lab-on-disc platform in clinical samples
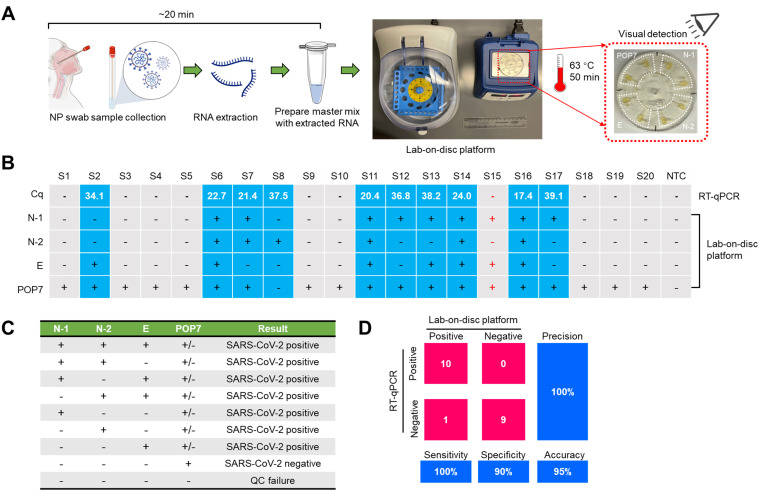
We conducted clinical validation of the lab-on-disc platform by testing 20 de-identified clinical NP swab samples and extracting their RNA by using commercial RNA extraction kits. Fig. 5A shows the workflow of lab-on-disc assay for clinical sample testing. The entire procedure can be completed in about 70 min from sample to answer. In the future, this test time could be further narrowed by interfacing the lab-on-disc platform with extraction-free detection strategies [35].
Fig. 5.
Validation of the lab-on-disc platform for multiplexed molecular detection of SARS-CoV-2 using 20 clinical NP swab samples (S1-S20). (A) Workflow and test time of the lab-on-disc platform. (B) Comparison of detection results of the lab-on-disc platform and RT-qPCR assays. + , positive; -, negative. S15 was marked red to shown the inconsistent result between the two approaches. According to the CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel, samples with Cq values below 40 were defined as positive. (C) Table summarizing results for the lab-on-disc platform. (D) Confusion matrix describing the overall performances of the lab-on-disc platform and the RT-qPCR assays between positive and negative samples. The RT-qPCR results are considered as the standard.
We confirmed that the 20 clinical samples included 10 SARS-CoV-2-positive and 10 SARS-CoV-2-negative samples by using CDC-released RT-qPCR assays. As shown in Fig. 5B, the positive Cq values ranged from 17.4 to 39.1. By testing the same samples, our lab-on-disc-based multiplexed SARS-CoV-2 detection approach presented with similar results, with the exception of S15 (Fig. 5B and S6). Based on the lab-on-a-disc detection, S15 is positive, as the N-1, E, and POP7 genes all produced positive results. The inconsistency between the assays may be attributed to RNA damage happening in the amplification region during sample preparation in the RT-qPCR assay. The results of the lab-on-disc platform are displayed in Fig. 5C. The POP7 internal control testing is important to classify the true negative and the quality control (QC) failures. If at least one of the three target genes of N-1, N-2, and E presents with positive signals, the result should be considered SARS-CoV-2 positive. Towards point-of-care diagnostics, we note that this lab-on-disc testing approach serves as a preliminary screening, especially for samples indicating positive values for only one target gene. Taking the RT-qPCR results as the standard, the confusion matrix indicates that our lab-on-disc platform has 100% sensitivity, 90% specificity, and 95% accuracy (Fig. 5D). Therefore, our monolithic, 3D-printed lab-on-disc platform is sensitive and reliable for COVID-19 diagnostics at the point of care.
4. Conclusions
In conclusion, we developed a monolithic, 3D-printed lab-on-disc platform for rapid, visual, multiplexed molecular detection of SARS-CoV-2. Unlike previously reported lab-on-disc systems [14], [15], our disc is fabricated in one step by 3D printing, eliminating the aligning and binding processes needed for multiple layers. Our lab-on-disc platform is able to simultaneously detect three different SARS-CoV-2 gene regions and one internal control gene region, enabling sensitive, reliable, and multiplexed molecular diagnostics. If needed, more reaction chambers can be integrated into single lab-on-disc platform for high-throughput screening of COVID-19. Within a 50-min incubation period, the lab-on-disc system has a high sensitivity (100 copies per chip) and high specificity (no cross-reactivity). Further, we clinically validated the lab-on-disc platform by testing 20 NP swab samples. Compared to the TaqMan probe-based RT-qPCR assays, our lab-on-disc platform demonstrated a sensitivity of 100% and an accuracy of 95%. Therefore, towards point-of-care diagnostics, our lab-on-disc platform provides simple, rapid, disposable, sensitive, reliable, and multiplexed SARS-CoV-2 detection and allows for preliminary screening for rapid COVID-19 diagnostics. In addition, by replacing the primers and targets, our platform could be easily applied to detect other infectious diseases, such as the influenza A virus, human immunodeficiency virus (HIV), and Zika virus. These advantages notwithstanding, our platform can be further improved in terms of sample preparation and throughput in the future. For example, through integrating on-chip nucleic acid extraction unit, we can develop a fully integrated, “sample-in-answer-out”, lab-on-disc detection system.
CRediT authorship contribution statement
Xiong Ding: Conceptualization, Methodology, Investigation, Data collection, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Ziyue Li: Investigation, Data collection, Formal analysis, Writing – review & editing. Changchun Liu: Conceptualization, Methodology, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published.
Declaration of Competing Interest
We confirm that we have no known competing financial interests or personal relationships.
Acknowledgement
This work was supported, in part, by the National Institutes of Health, United States (R01EB023607, R61AI154642, and R01CA214072).
Biographies
Xiong Ding is a postdoctoral researcher in the Department of Biomedical Engineering at the University of Connecticut Health Center, USA. He obtained his PhD degree in Biochemistry and Molecular Biology from Zhejiang University, China, in 2017. His research is dedicated to the fields of biomedical molecular diagnostics and biosensors.
Ziyue Li is currently a PhD student of Biomedical Engineering at the University of Connecticut, USA. He received his Master’s degree in Electronics Engineering from the University of Chinese Academy of Sciences and Bachelor’s degree in Electrical Engineering from Nankai University in China. His research focuses on microfluidic chips, biosensors, and point-of-care diagnostics.
Changchun Liu is currently an Associate Professor of Biomedical Engineering at the University of Connecticut Health Center, USA. He received his PhD degree in Electronics Engineering from the Institute of Electronics, Chinese Academy of Sciences, Beijing, China. His research focuses on microfluidic chips, biosensors, and CRISPR technology for point-of-care diagnostics.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2021.130998.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.WHO, WHO Coronavirus (COVID-19) Dashboard, https://covid19whoint/, (2021).
- 2.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furukawa N.W., Brooks J.T., Sobel J. Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.201595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. Jama. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M., Meshorer E., Benedek G., Fogel I., Oiknine-Djian E., Gertler A., Rotstein Z., Lavi B., Dor Y., Wolf D.G., Salton M., Drier Y., Hebrew T. University-Hadassah COVID- Diagnosis, large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin. Microbiol. Infect. 2020;26:1248–1253. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta. 2020;507:139–142. doi: 10.1016/j.cca.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., Deconinck L., Lescure F.X., Lucet J.C., Bouadma L., Timsit J.F., Descamps D., Yazdanpanah Y., Casalino E., Houhou-Fidouh N., Emergency Department Influenza Study G. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J. Clin. Microbiol. 2020;58:e00630–20. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian J., Boswell S.A., Chidley C., Lu Z.-X., Pettit M.E., Gaudio B.L., et al. Enhanc. isothermal Amplif. Assay. Viral Detect., Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X., Yin K., Li Z., Sfeir M.M., Liu C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens. Bioelectron. 2021;184 doi: 10.1016/j.bios.2021.113218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Tanner N.A. Development of multiplexed reverse-transcription loop-mediated isothermal amplification for detection of SARS-CoV-2 and influenza viral RNA. Biotechniques. 2021;70:167–174. doi: 10.2144/btn-2020-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang W.S., Lim D.H., Yoon J., Kim A., Lim M., Nam J., Yanagihara R., Ryu S.W., Jung B.K., Ryoo N.H., Lim C.S. Development of a multiplex loop-mediated isothermal amplification (LAMP) assay for on-site diagnosis of SARS CoV-2. PloS One. 2021;16 doi: 10.1371/journal.pone.0248042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding S., Chen G., Wei Y., Dong J., Du F., Cui X., Huang X., Tang Z. Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang M., Wang G., Kong S.-K., Ho H.-P. A review of biomedical centrifugal microfluidic platforms. Micromachines. 2016;7:26. doi: 10.3390/mi7020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strohmeier O., Keller M., Schwemmer F., Zehnle S., Mark D., von Stetten F., Zengerle R., Paust N. Centrifugal microfluidic platforms: advanced unit operations and applications. Chem. Soc. Rev. 2015;44:6187–6229. doi: 10.1039/c4cs00371c. [DOI] [PubMed] [Google Scholar]
- 16.Sayad A., Ibrahim F., Uddin S.M., Cho J., Madou M., Thong K.L. A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform. Biosens. Bioelectron. 2018;100:96–104. doi: 10.1016/j.bios.2017.08.060. [DOI] [PubMed] [Google Scholar]
- 17.Maguire I., O’Kennedy R., Ducrée J., Regan F. A review of centrifugal microfluidics in environmental monitoring. Anal. Methods. 2018;10:1497–1515. [Google Scholar]
- 18.Miyazaki C.M., Carthy E., Kinahan D.J. Biosensing on the centrifugal microfluidic lab-on-a-disc platform. Processes. 2020;8:1360. [Google Scholar]
- 19.Palenzuela C.L.M., Pumera M. (Bio) Analytical chemistry enabled by 3D printing: sensors and biosensors. TrAC Trends Anal. Chem. 2018;103:110–118. [Google Scholar]
- 20.Ho C.M.B., Ng S.H., Li K.H.H., Yoon Y.-J. 3D printed microfluidics for biological applications. Lab a Chip. 2015;15:3627–3637. doi: 10.1039/c5lc00685f. [DOI] [PubMed] [Google Scholar]
- 21.Ding X., Xu Z., Yin K., Sfeir M., Liu C. Dual-Priming isothermal amplification (DAMP) for highly sensitive and specific molecular detection with ultralow nonspecific signals. Anal. Chem. 2019;91:12852–12858. doi: 10.1021/acs.analchem.9b02582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC, CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel, https://wwwfdagov/media/134922/download, (2020). [DOI] [PMC free article] [PubMed]
- 23.Focke M., Stumpf F., Faltin B., Reith P., Bamarni D., Wadle S., Müller C., Reinecke H., Schrenzel J., Francois P., Mark D., Roth G., Zengerle R., von Stetten F. Microstructuring of polymer films for sensitive genotyping by real-time PCR on a centrifugal microfluidic platform. Lab a Chip. 2010;10:2519–2526. doi: 10.1039/c004954a. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Tian F., Liu C., Feng Q., Ma T., Zhao Z., Li T., Jiang X., Sun J. Hand-powered centrifugal microfluidic platform inspired by the spinning top for sample-to-answer diagnostics of nucleic acids. Lab a Chip. 2018;18:610–619. doi: 10.1039/c7lc01234a. [DOI] [PubMed] [Google Scholar]
- 25.Yin K., Ding X., Xu Z., Li Z., Wang X., Zhao H., Otis C., Li B., Liu C. Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip. Sens. Actuators B: Chem. 2021;344 doi: 10.1016/j.snb.2021.130242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin K., Ding X., Li Z., Sfeir M.M., Ballesteros E., Liu C. Autonomous lab-on-paper for multiplexed, CRISPR-based diagnostics of SARS-CoV-2. Lab a Chip. 2021 doi: 10.1039/d1lc00293g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Commentary: Middle east respiratory syndrome coronavirus (mers-cov): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Peiris J.S. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D.X., Liang J.Q., Fung T.S. Human coronavirus-229E,-OC43,-NL63, and-HKU1 (Coronaviridae) Encycl. Virol. 2021:428–440. [Google Scholar]
- 30.Mitani Y., Lezhava A., Kawai Y., Kikuchi T., Oguchi-Katayama A., Kogo Y., Itoh M., Miyagi T., Takakura H., Hoshi K., Kato C., Arakawa T., Shibata K., Fukui K., Masui R., Kuramitsu S., Kiyotani K., Chalk A., Tsunekawa K., Murakami M., Kamataki T., Oka T., Shimada H., Cizdziel P.E., Hayashizaki Y. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat. Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 31.Higgins O., Smith T.J. Loop-primer endonuclease cleavage–loop-mediated isothermal amplification technology for multiplex pathogen detection and single-nucleotide polymorphism identification. J. Mol. Diagn. 2020;22:640–651. doi: 10.1016/j.jmoldx.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Malpartida-Cardenas K., Rodriguez-Manzano J., Yu L.-S., Delves M.J., Nguon C., Chotivanich K., Baum J., Georgiou P. Allele-specific isothermal amplification method using unmodified self-stabilizing competitive primers. Anal. Chem. 2018;90:11972–11980. doi: 10.1021/acs.analchem.8b02416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varona M., Anderson J.L. Visual detection of single-nucleotide polymorphisms using molecular beacon loop-mediated isothermal amplification with centrifuge-free DNA extraction. Anal. Chem. 2019;91:6991–6995. doi: 10.1021/acs.analchem.9b01762. [DOI] [PubMed] [Google Scholar]
- 34.Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalli M.A., Langmade J.S., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.