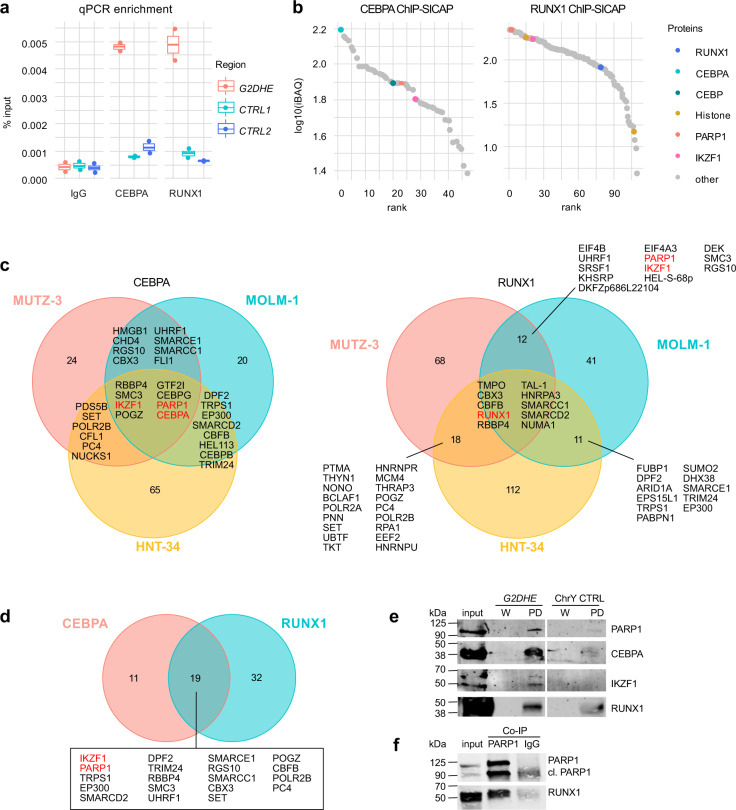
Fig. 2. CEBPA and RUNX1 form a complex with IKZF1 and PARP1 at G2DHE in 3q-rearranged AML.
ChIP-SICAP was performed in three different 3q-rearranged cell lines using antibodies against CEBPA and RUNX1 or an unspecific IgG control. Experiments were carried out in duplicates (n = 2). a, b show exemplary results for the cell line MUTZ-3. a The DNA fraction of the ChIP-SICAP experiment was used for enrichment quantification of the G2DHE region by qPCR compared to two unrelated control regions (CTRL). b The protein fraction of the ChIP-SICAP experiments was analyzed by mass spectrometry to identify chromatin-bound interactors of RUNX1 and CEBPA. Proteins enriched in the CEBPA- or RUNX1-captured samples over the IgG controls in both MUTZ-3 replicates were ranked according to their iBAQ intensity. c Overlap of the proteins identified in different cell lines. d Overlap of proteins that were identified in at least two cell lines per bait. e DNA-streptavidin pull-down (PD) of CEBPA, RUNX1, IKZF1, and PARP1. Nuclear lysate of MUTZ-3 cells was incubated with biotinylated DNA probes containing the G2DHE sequence or a deserted control region from chromosome Y (ChrY CTRL). Proteins associated with the DNA probes were purified and analyzed by western blot using antibodies against CEBPA, RUNX1, IKZF1, and PARP1. Input and an aliquot of the last wash steps (W) served as control for the PD samples. f Co-immunoprecipitation of PARP1 and RUNX1. Total cell lysate of MUTZ-3 was used for immunoprecipitation either with a PARP1 antibody or a control IgG. 75 µg of lysate was used as input control. Western blot was performed using antibodies directed against PARP1 and RUNX1.