Figure 2.
Sex and APOE genotype drive amyloid pathology and microglial gene expression profiles in APOE-FAD mice
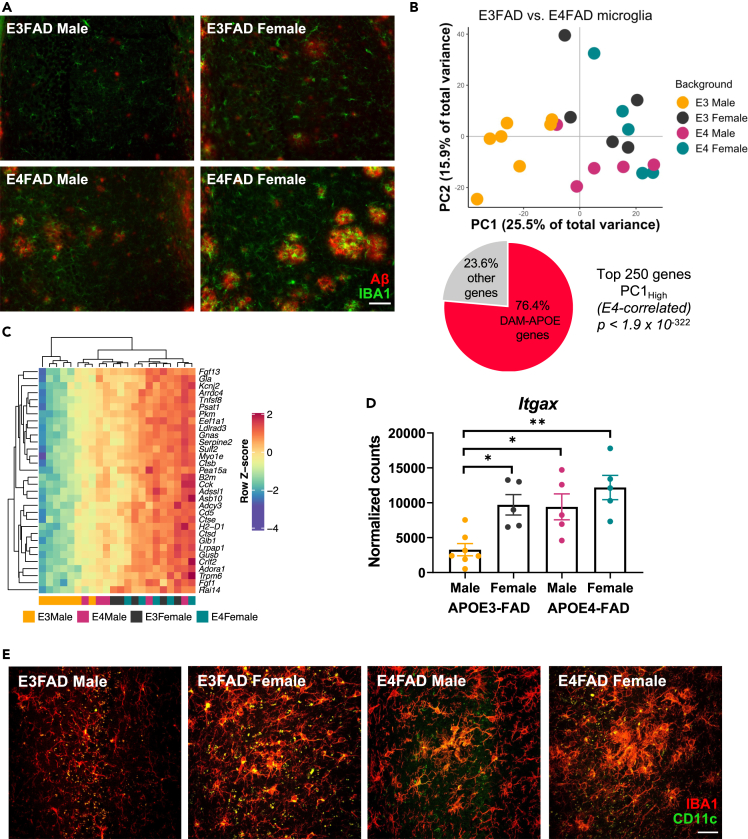
(A) Representative images of immunohistochemical staining for amyloid-β (red) and the microglial marker IBA1 (green) in male and female APOE3- and APOE4-FAD mice. Scale bar, 50 μm.
(B) Samples separate by APOE and sex along PC1, with APOE3-FAD males clustering separately from mice of both sexes and APOE3-FAD females. A large proportion of the genes driving this separation (76.4%) are DAM-APOE genes. APOE3-FAD males are shown in yellow, APOE3-FAD females in gray, APOE4-FAD males in magenta, and APOE4-FAD females in teal.
(C) Heatmap of the DAM-APOE genes associated with sample clustering along PC1. Gene expression levels were normalized, scaled, and centered and are displayed as Z scores.
(D and E) Levels of the DAM-APOE marker, CD11c, are increased in APOE3-FAD females and in APOE4-FAD mice of both sexes, both at the (D) transcript and (E) protein levels. (D) Gene expression of Itgax in normalized counts; data are presented as mean (±SEM) values; n = 5–7/group. ∗p < 0.05, ∗∗p < 0.01; one-way ANOVA with Tukey’s multiple comparisons test correction. (E) Representative immunofluorescence images of microglia (IBA1, red) and CD11c (green) in the hippocampal subiculum of male and female APOE3- and APOE4-FAD mice. Scale bar, 25 μm.