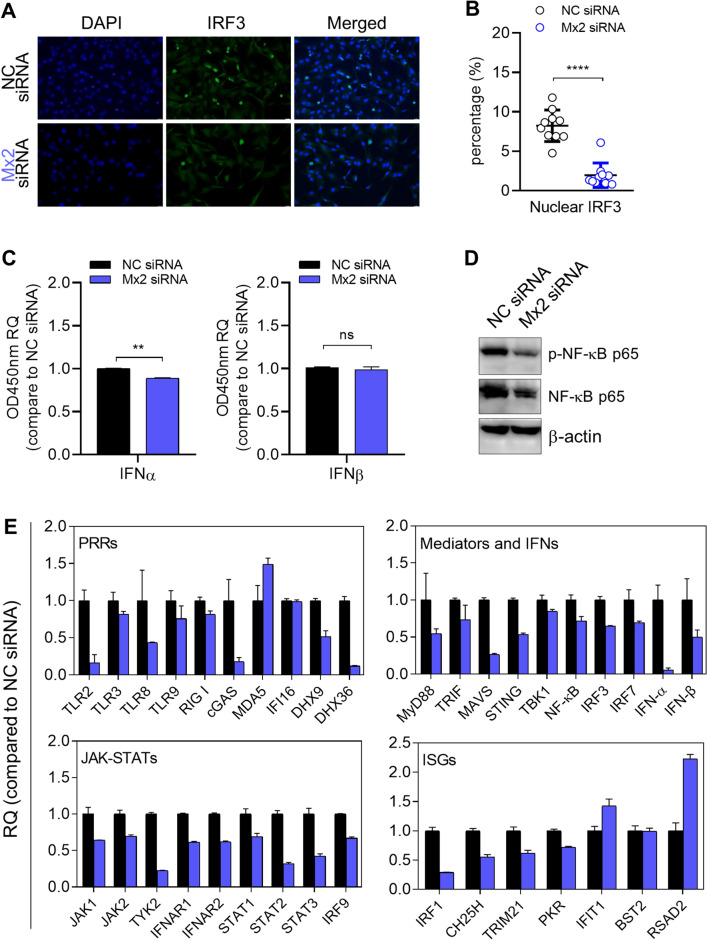
Figure 4.
The effects of Mx2 depletion on antiviral innate response and intrinsic mRNA accumulation of innate immune genes in resistant A172 cells. (A) Investigation of nuclear IRF3 in A172 cells via immunofluorescence staining. (B) Quantification of IRF3 localized in the A172 nucleus by automatic counting using Image J software. 10–15 fields of each slide were randomly chosen for counting the numbers of IRF3-DAPI coincident dots. (C) ELISA quantification of secreted IFNα and IFNβ in cell supernatant. RQ: relative quantity, values that correspond to optical densities at OD 450 nm compared to values in NC siRNA-transfected A172 cells. (*p < 0.05; **p < 0.01; compared with NC siRNA-treated A172 cells. Error bars represent SD.) (D) Expression of effector proteins associated with the NF-κB signaling pathway. Blots were cut and probed with indicated antibodies, respectively. β-actin serves as a loading control. (E) mRNA accumulation of indicated immune genes in Mx2-depleted A172 cells. Mx2-depleted or mock-depleted A172 cells were harvested at 24 h post-transfection. Total RNA was isolated and analyzed by qPCR assay for mRNA levels of indicated immune genes. Used primers are shown in Table 3. RQ: relative quantity, compared to transcript levels in mock-depleted A172 cells. Bar, mean ± SD. GAPDH serves as a normalization control.