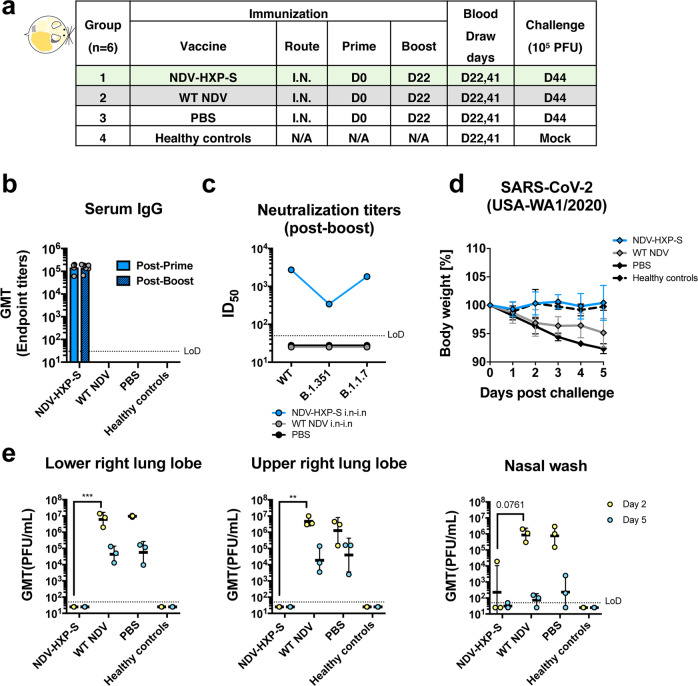
Fig. 5. Live NDV-HXP-S via the intranasal route induces protective antibody responses in hamsters.
a Design of the study. Eighteen-to-twenty-week-old female Golden Syrian hamsters were used. Group 1 (n = 6) was vaccinated with 106 EID50 of live NDV-HXP-S. Group 2 (n = 6) was vaccinated with 106 EID50 of live WT NDV as the vector-only control. Group 3 (n = 6) was vaccinated with PBS as the negative control. Group 4 (n = 6) were the healthy controls. The vaccine was administered via the intranasal (I.N.) route at D0 and D22. Blood was collected at D22 and D41. Group 1–3 were challenged with 105 PFU of the USA-WA1/2020 strain at D44. Group 4 was mock-challenged with PBS. b Spike-specific serum IgG. Antibodies in post-prime (D22, solid blue bar) and post-boost (D41, pattern blue bar) sera were measured by ELISAs. GMT endpoint titers were graphed. The error bars represent geometric SD. c Neutralizing activity of serum antibodies. Post-boost sera from groups 1–3 were pooled within each group and tested in neutralization assay against USA-WA/2020 strain (WT), B.1.351 variant, and B 1.1.7 variant in technical duplicate. Serum dilutions inhibiting 50% of the infection (ID50) were plotted (LoD = 1:50; An ID50 = 1:25 was assigned to negative samples). d Body weight change of hamsters. Bodyweight was recorded for 5 days after the challenge. The error bars represent geometric SD. (c and d, blue: NDV-HXP-S; gray: WT NDV; black: PBS) e Viral load in the lungs and nasal washes. The lower right and upper right lung lobes of a subset of animals (n = 3) from each group were collected at day 2 (yellow) and day 5 (blue) post challenge. Each lung lobe was homogenized in 1 mL PBS. Nasal washes were collected in 0.4 mL of PBS. Viral titers were measured by plaque assay on Vero E6 cells and plotted as GMT of PFU/mL (LoD = 50 PFU/mL; a titer of 25 PFU/mL was assigned to negative samples). The error bars represent geometric SD. Statistical difference was analyzed by two-way ANOVA corrected for Dunnett’s multiple comparisons test (**p < 0.0017; ***p = 0.0009).