Figure 2. PER2 stability is tightly regulated by post-translational modifications.
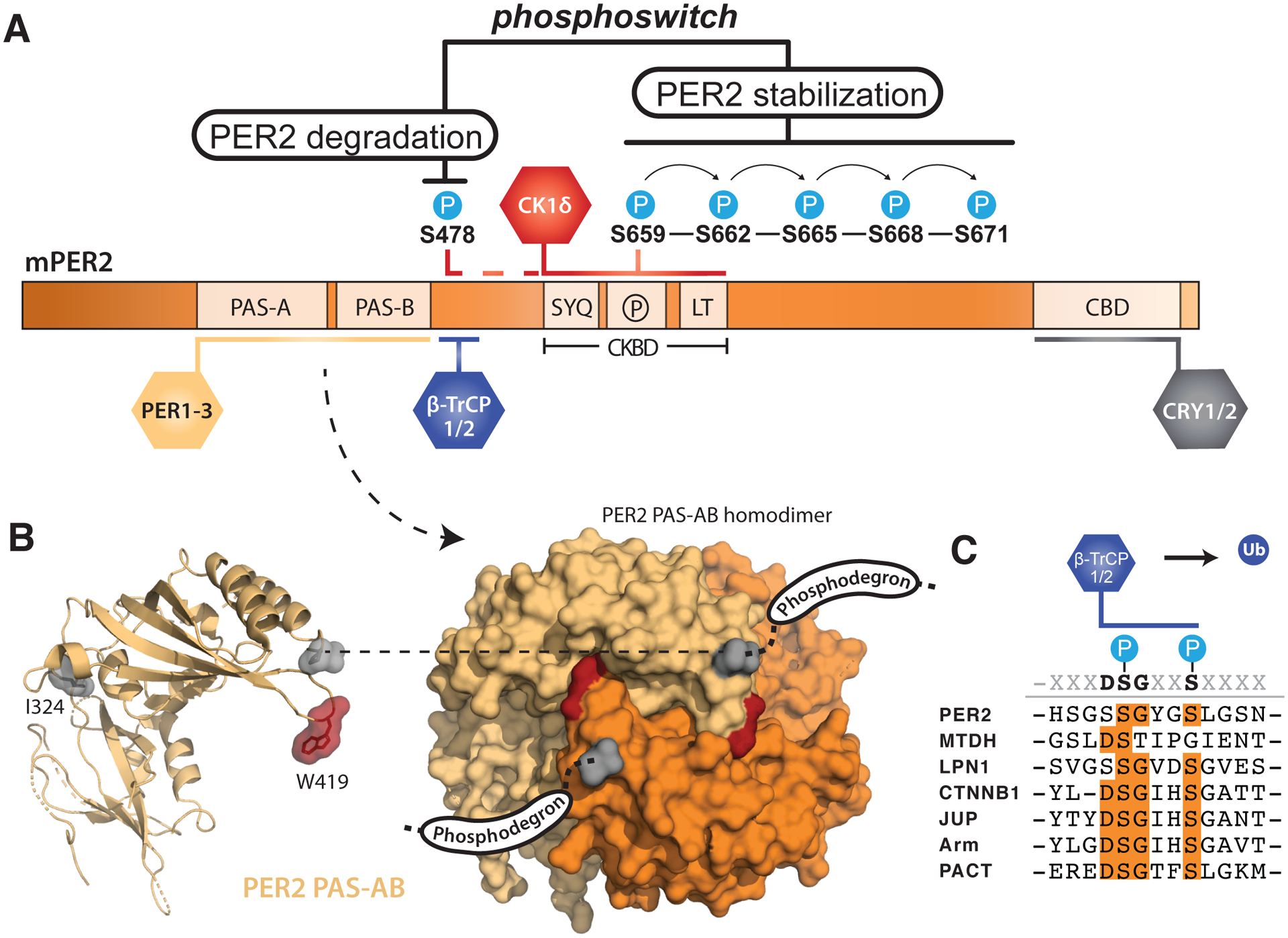
(A) Domain map of mouse PER2 depicting clock protein binding sites and the CK1δ-dependent phosphoswitch model. CK1δ-dependent phosphorylation of the FASP region (P) within the CKBD antagonizes activity at the upstream phosphodegron, which is used to recruit the E3 ubiquitin ligases, β-TrCP1/2, for subsequent proteasomal degradation of PER2. (B) Left, cartoon representation of the mouse PER2 PAS-AB domain monomer with the location of the Edo mutation (I324N) and Trp residue required for dimerization (W419) highlighted, PDB: 3GDI. Right, surface representation of the PER2 PAS-AB homodimer. The phosphodegron is located in a disordered region immediately downstream of the PAS-B domain; the C-terminal residue is depicted in surface mode (gray) to show how each respective phosphodegron is poised to protrude from the same face of the dimer. (C) Alignment of phosphodegrons within human proteins that are targeted by β-TrCP1/2.
