Figure 3. Regulatory mechanisms control CK1δ activity in the mammalian clock.
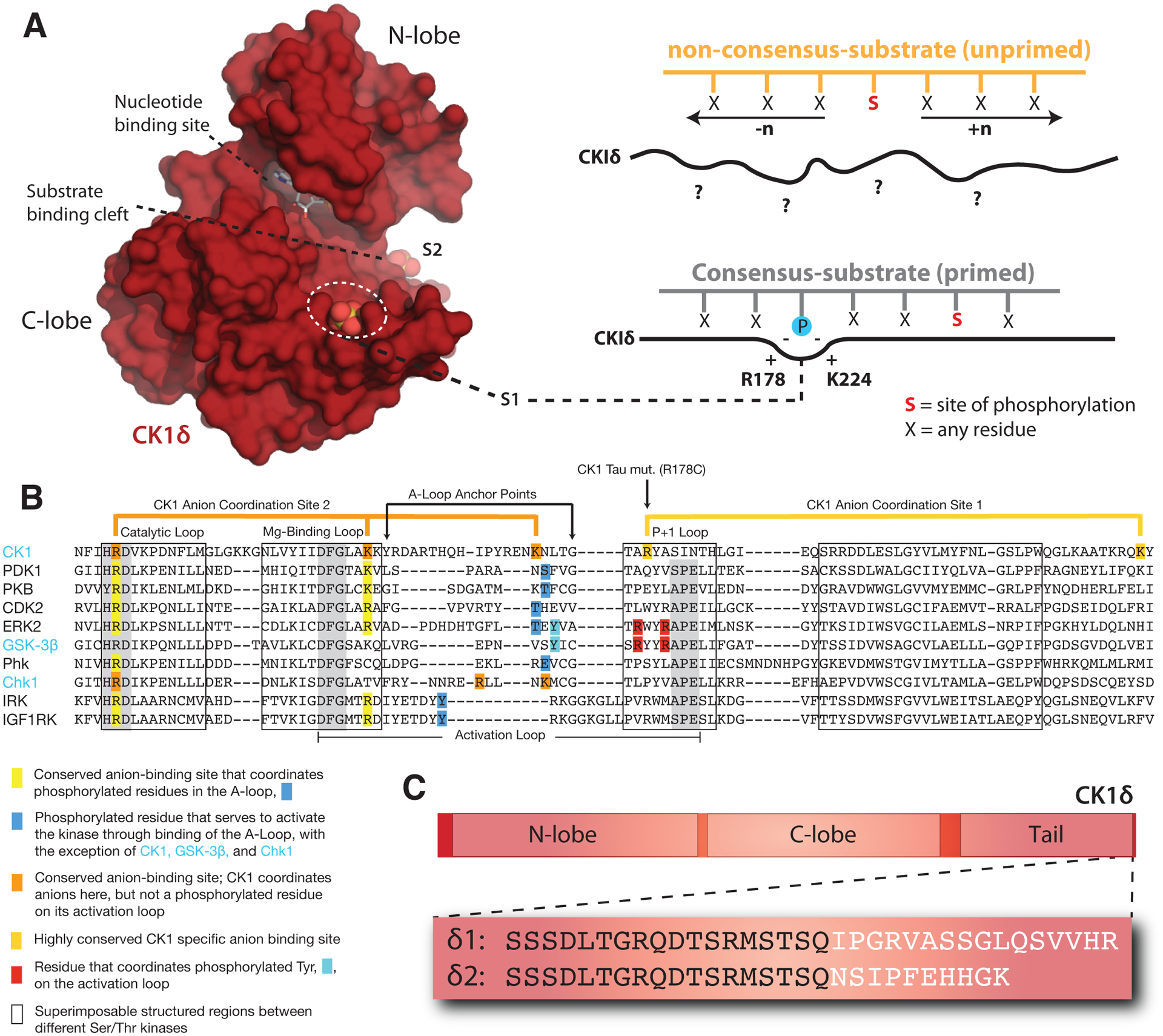
(A) Left, Surface representation of the CK1δ kinase domain, PDB: 5×17. The substrate binding cleft is flanked by two highly conserved anion binding sites, S1 and S2. Right, CK1 is thought to use S1 to bind phosphorylated (or ‘primed’) substrates, leading to phosphorylation of the CK1 consensus motif, pSxxS. CK1 also exhibits non-consensus activity on unprimed sites. (B) Alignment of the activation loop and nearby regions in representative Ser/Thr kinases. CK1 lacks the conserved APE motif in the P+1 loop involved in substrate recognition. (C) Differences between the alternatively spliced variants CK1δ1 and CK1δ2 at the C-terminus of the kinase.
