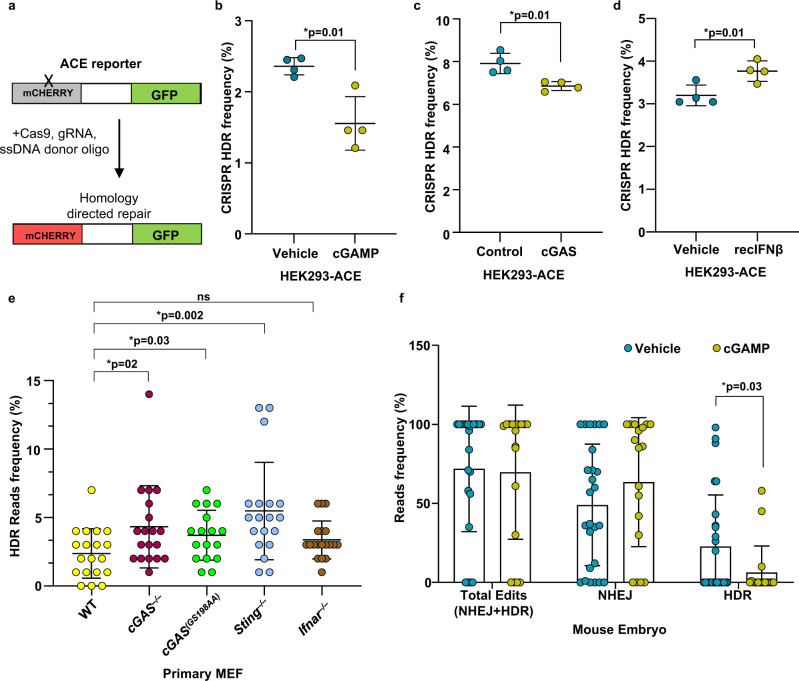
Fig. 8. cGAMP suppresses CRISPR-Cas9 genome editing.
a GFP-positive HEK293-ACE CRISPR/Cas9 reporter cells were transfected with recombinant Cas9, gRNA, and donor template to repair DNA sequences encoding mutant non-fluorescent mCherry to functional fluorescent mCherry expression cassettes. b The percentages of HEK293-ACE CRISPR/Cas9 reporter cells mock stimulated and stimulated with cGAMP determined to be mCherry positive are presented (n = cell culture replicates, data presented are mean ± s.d.; two-tailed unpaired t test; *p < 0.05 indicates significance compared to respective groups; ns indicates not significant). c The percentages of HEK293-ACE cells stably expressing cGAS and control (empty plasmid) determined by flow cytometry to be mCherry positive are presented (data presented are mean ± s.d., n = 4 cell culture replicates, data presented are mean ± s.d.; two-tailed unpaired t test; *p < 0.05 indicates significance compared to respective groups; ns indicates not significant). d GFP-positive HEK293-ACE CRISPR/Cas9 reporter cells were treated with human recombinant interferon-β (50 ng/ml) and then transfected with recombinant Cas9, gRNA, and donor template to repair DNA sequences encoding mutant non-fluorescent mCherry to functional fluorescent mCherry expression cassettes, followed by flow cytometry. Quantification of flow cytometry data is presented (n = 5 cell culture replicate; data presented are mean ± s.d.; two-tailed unpaired t test; *p < 0.05 indicates significance compared to respective groups; ns indicates not significant). e The Rosa26 locus was edited using CRISPR/Cas9 in WT, cGAS−/−, cGAS(GS198AA), Sting−/−, and Ifnar−/− mouse primary embryonic fibroblasts and subsequently PCR amplified for next-generation sequencing and CRISPResso analysis. The frequency of CRISPR/Cas9-mediated homology-directed repair outcomes in these genotypes are presented (N = 19 for WT, N = 19 for cGAS−/−, N = 17 for cGAS(GS198AA), N = 19 for Sting−/−, and N = 19 for Ifnar−/− cell culture replicates; data presented are mean ± s.d.; two-tailed unpaired t test, no adjustments were made for multiple comparisons; *p < 0.05 indicates significance compared to respective groups; ns indicates not significant). Each data point represents percentage of sequence reads, indicative of HDR, n = 19 cell culture replicates for each genotype. f The frequency of CRISPR/Cas9-mediated genome-editing outcomes in mouse embryos in the presence or absence of cGAMP was determined as described in the schematic (Supplementary Fig. 10c; n = 26 embryos for vehicle, n = 18 embryos for cGAMP; data presented are mean ± s.d.; two-tailed unpaired t test; *p < 0.05 indicates significance compared to respective groups; ns indicates not significant). Each data point represents percentage of sequence reads within the indicated outcomes (Total edits, NHEJ, HDR).