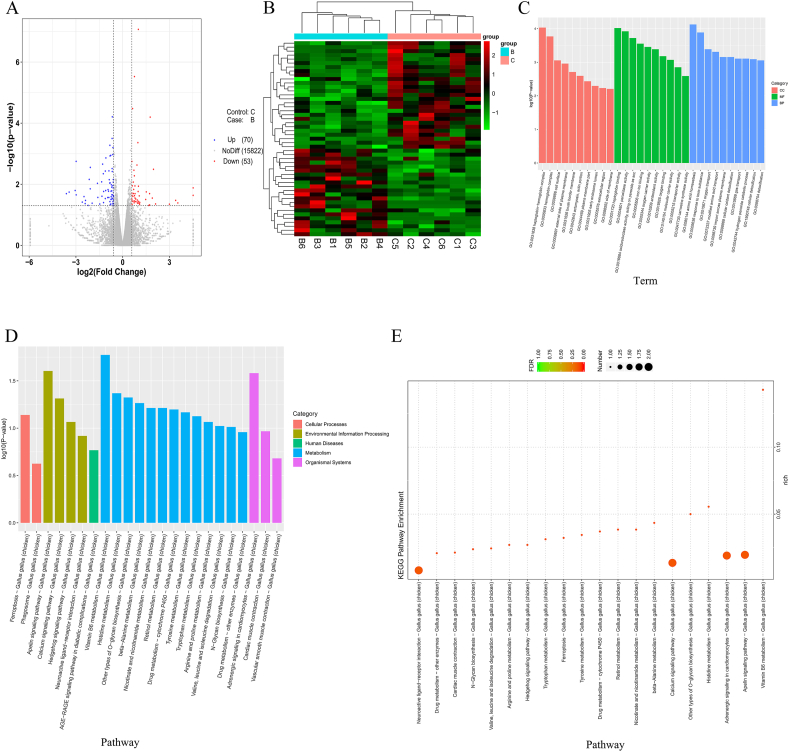
Fig. 2.
The RNA-seq of chicken cecal epithelial cells. (A) Volcano plot of differentially expressed genes. (B) Clustering heatmap of differentially expressed genes. Each row represents a gene, and each column is a sample. The red color indicates highly expressed genes, and the green color indicates genes showing with low expression. B1 to B6 refer to Group B, the Clostridium butyricum group; C1 to C6 refer to Group C, control group. (C) Histogram of the Gene Ontology (GO) enrichment analysis results. The horizontal coordinate is the term of the GO level 2 term, the vertical coordinate is the -log10(P-value) showing the enrichment of each term, and the number on the column shows the number of differential genes enriched in each term. In each GO classification, the top 10 GO terms with the lowest P-value, i.e., the most significant enrichment, were selected for display. (D and E) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis results. According to the KEGG enrichment analysis of the differentially expressed genes, the first 20 pathways with the smallest P-value, i.e., the most significant enrichment, were selected for display in D. The abscissa is the name of the pathway, and the ordinate is the -log10(P-value) showing the enrichment of each pathway; the number of genes in the column shows the number of differentially expressed genes enriched in the pathway (E).