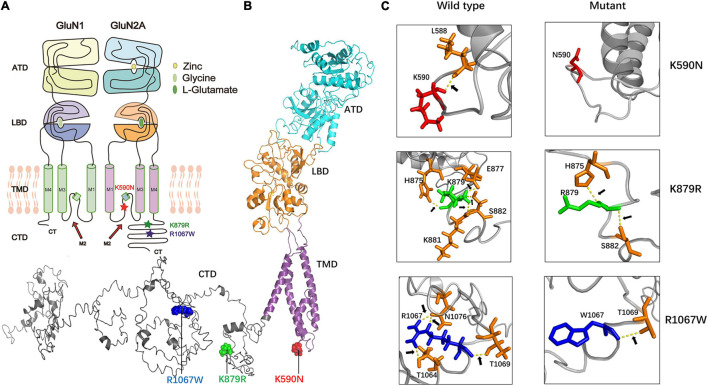
FIGURE 2.
The alterations of hydrogen bonds with surrounding amino acids. (A) The locations of missense mutations in topological structure of GluN1/GluN2A. Residue K590N in GluN2A lies within the channel pore of the NMDAR, while residues K879R and R1067W lie in carboxyl-terminal domain. Mutation K590N was colored in red, mutation K876R was colored in green, and mutation R1067W was colored in blue. (B) Schematic illustration of the location of mutations in the three-dimensional structure of GluN2A. (C) Alterations of hydrogen bonds with surrounding amino acids. In wild type, residue K590 forms one hydrogen bond with L588. In the mutant, this hydrogen bond was destroyed. In wild type, residue K879 forms hydrogen bonds with H875, E877, K881, and S882 while in the mutant, the hydrogen bonds with E877 and K881 were destroyed. In wild type, residue R1067 forms hydrogen bonds with T1064, T1069, and N1076 while in the mutant, only hydrogen bond with T1069 was kept.