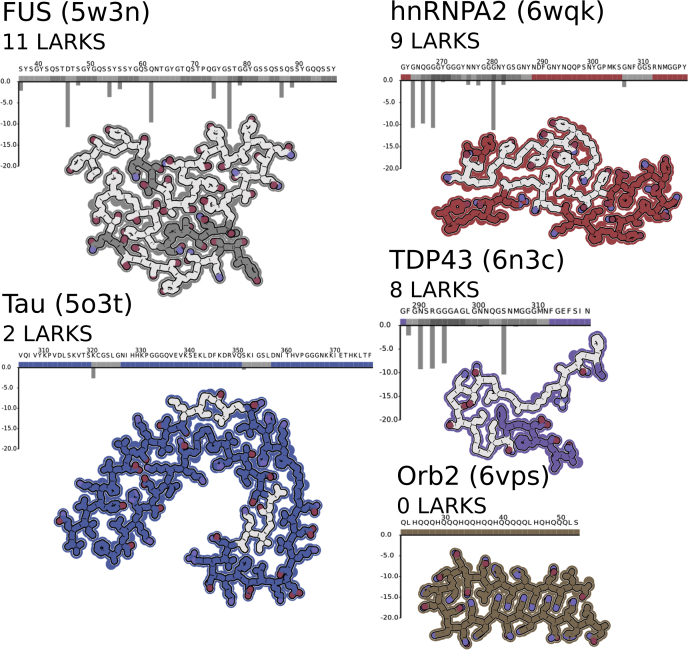
Figure 4.
LARKS alter amyloid structure. All images are made from a single chain and a single layer of the fibril, being viewed down the fibril axis. LARKS predictions and sequences are shown above structures. Gray boxes above the sequence indicate predicted LARKS, and the corresponding residues in the protein images have a gray interior. There are three structures of LARKS-rich LCDs: from FUS, hnRNPA2, and TDP43 (Protein Data Bank [PDB] IDs: 5W3N, 6WQK, and 6N3C). And two structures from irreversible amyloid Tau and Orb2. Tau is associated with Alzheimer's pathogenesis, and the presented structure (PDB ID: 5O3T) was made by seeding purified protein with extracts from the brain of a deceased patient with Alzheimer's disease. Orb2 (PDB ID: 6VPS) from Drosophila melanogaster forms stable amyloid fibrils associated with memory formation in flies and was purified from fly brains. Qualitatively, the structures of proteins associated with dynamic MLOs (FUS, hnRNPA2, and TDP43) have more LARKS and have more kinks in their backbones when compared with the stable amyloids—disease associated (Tau) or functional amyloid (Orb2). LARKS, low-complexity amyloid-like reversible kinked segment; LCD, low-complexity domain.