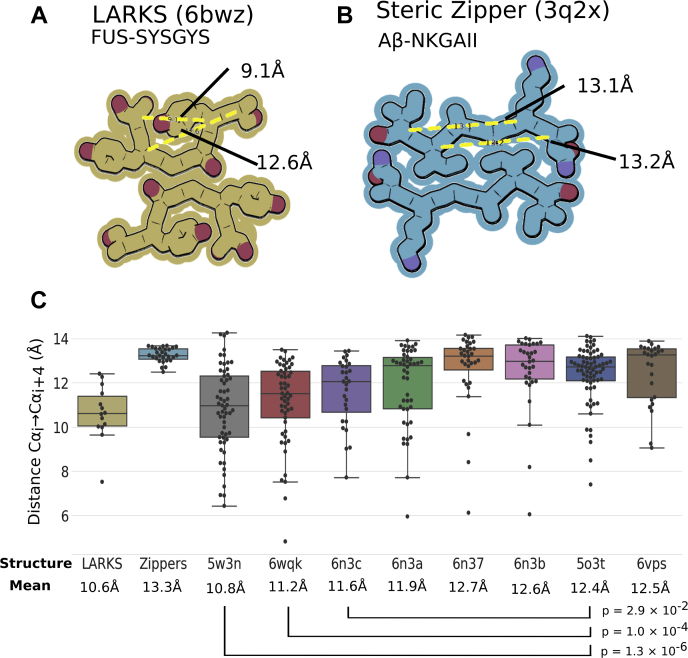
Figure 5.
LARKS in protein structure. A, example of a LARKS structure, FUS-SYSGYS, shows the kinked backbone and minimal interface between mating sheets. Measuring all possible Cαi → Cαi + 4 pairs finds distances of 12.6 and 9.1 Å, a result of the kinked backbone. B, an example of a steric zipper, Aβ-NKGAII, to show the extensive mating interface between the extended and pleated β-sheets. Cαi → Cαi + 4 distances are 13.1 and 13.2 Å because the regular spacing from the β-sheet structure. C, box and whisker plots of Cαi → Cαi + 4 distances. Each black point is a single distance measurement from either a sample of LARKS crystal structures, steric zipper structures, or the full-length proteins shown in Figure 4 as well as additional TDP43 structures in Fig. S2. The LARKS-rich LCD amyloids (FUS-5w3n, hnRNPA2-6wqk, and TDP43-6n3c) have significantly closer Cαi → Cαi + 4 distances than found in irreversible amyloids (Tau-5o3t and Orb2-6vps) reflecting how LARKS affect protein structure by interrupting β-sheets. LARKS, low-complexity amyloid-like reversible kinked segment; LCD, low-complexity domain.