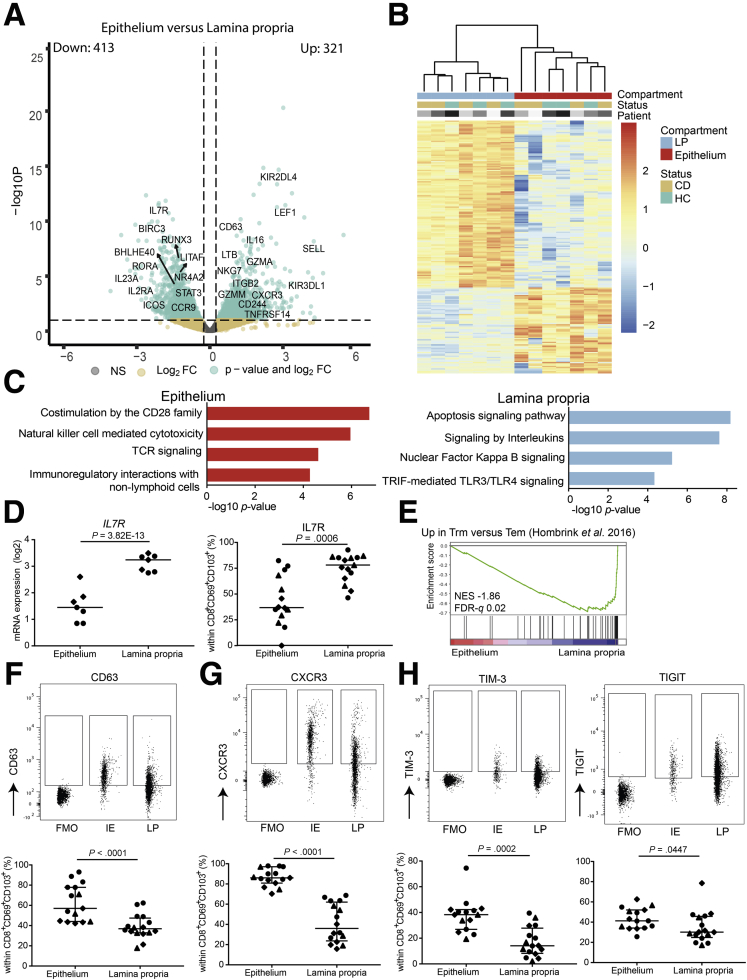
Figure 5.
Location shapes the profile of intestinal CD103+CD69+CD8+T cells. (A) Volcano plot of the expressed genes, with a nominal P value <0.99, comparing intraepithelial to lamina propria (LP) CD8+CD69+CD103+ T cells; selected genes are highlighted. On the x-axis, the log2 fold change (log2FC) is shown, and on the y-axis, the -log10 P value (-log10P) is shown. Gray indicates not significantly differentially expressed genes, yellow indicates genes with a log2FC >0.25 and -log10P >10 × 10–2.5, green indicates genes with a log2FC > 0.25 and -log10P < 10 × 10–2.5. (B) Heatmap of the top 200 differentially expressed genes comparing CD103+ intraepithelial and lamina propria T cells with hierarchical clustering on the columns concerning compartment, status, and patient. Rows are z score normalized. (C) Pathway terms related to the 321 genes upregulated in intraepithelial (top) and 413 genes upregulated in LP (bottom) CD8+CD69+CD103+ T cells. (D) Messenger RNA (mRNA) expression (log2 counts; right) and percentage (left) of ileal CD8+CD69+CD103+ T cells expressing IL7R (CD127) in healthy control subjects (n = 3–5; diamonds) and CD patients from inflamed (circles) and noninflamed (triangles) ileum (paired, n = 4–5). Comparison was performed with Wald’s statistic and a paired 2-tailed t test, respectively. (E) Gene set enrichment analysis of Trm genes in humans (identified by Hombrink et al)25 in pairwise comparisons involving intraepithelial and LP CD8+CD69+CD103+ T cells derived from the ileum of healthy adult control subjects and CD patients pooled, represented by the normalized enrichment score and FDR statistical value (FDRq). (F) Representative gating strategy including Fluorescence Minus One (FMO) control (upper panel) and quantification (lower panel) of CD63 in CD8+CD69+CD103+ T cells comparing epithelium (IE) and LP. Bars represent median and interquartile range. Comparison was performed with a paired 1-tailed test. Symbols as per panel D. Healthy control subjects: n = 7; CD patients: n = 3–6. (G) As per panel F but for CXCR3. (H) As per panel F but for TIM-3 (left) and TIGIT (right). Comparison was performed with a paired 2-tailed t test. NS, not significant.