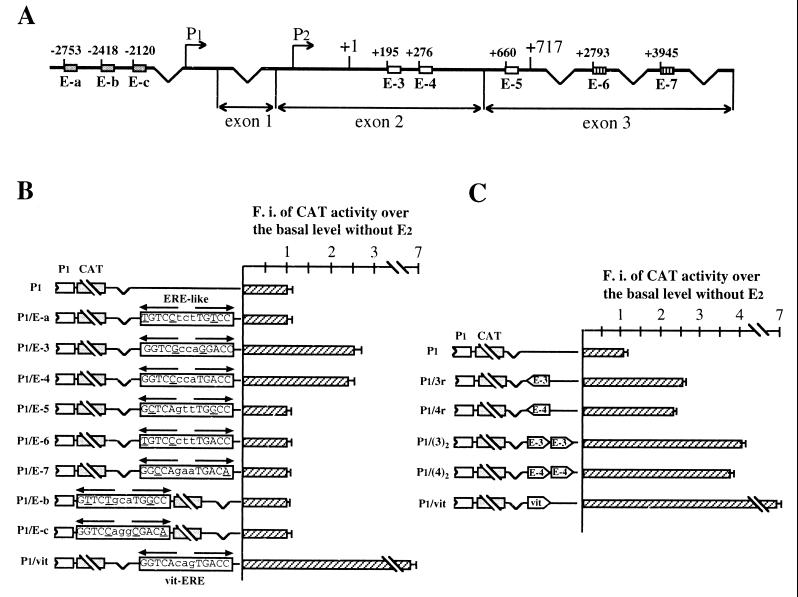
FIG. 3.
(A) Schematic structure of the human bcl-2 gene and location of putative EREs. Eight sequences have been identified: three in the 1.2-kb region 5′ to the P1 promoter (E-a to E-c, grey boxes), three within the 717-bp coding region (E-3 to E-5, white boxes), and two within the 3′ untranslated region (E-6 and E-7, boxes with vertical lines). The broken arrows indicate the 5′ ends of P1 and P2 promoters. Numbers on top of each box identify the position of the first nucleotide. Exons 2 and 3 are separated by a 225-kb intron. (B) Analysis of CAT activity measured in MCF-7 cells separately transfected with the constructs schematically represented on the left. Each construct contains one of the potential ERE-like sequences, cloned either 1.7 kb downstream from (P1/E-a and P1/E-3 to P1/E-7) or just 3′ to (P1/E-b and P1/E-c) the P1 promoter. P1 and P1/vit constructs were included as negative and positive controls, respectively. Opposite-pointing arrows on top of each sequence denote the ERE half-palindromes. Mutant nucleotides compared to consensus ERE are underlined. Insertion of E-3 and E-4 sequences confers hormone sensitivity on reporter constructs. (C) Estrogen responsiveness of CAT activity measured in MCF-7 cells transfected with the plasmids schematically represented on the left and containing, respectively, one copy of E-3 or E-4 sequence in reverse orientation (P1/3r and P1/4r) or two copies of each sequence in forward orientation, cloned as described for panel B. P1 and P1/vit constructs were also included as negative and positive controls, respectively. As for panel B, reporter gene activities are as detailed in the Fig. 2 legend; the error bars reflect the standard errors of three separate assays. Both E-3 and E-4 oligonucleotides behave as classical enhancer sequences. Transfections of 5- to 7-day hormone-starved MCF-7 cells were performed as described in Materials and Methods. F.i., fold induction.