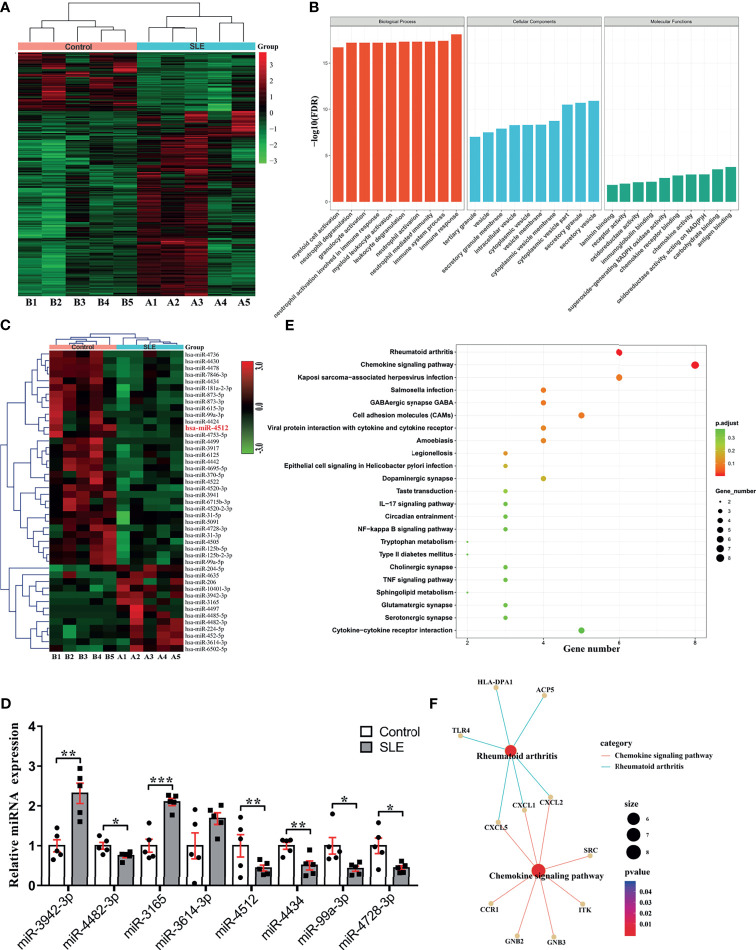
Figure 1.
miRNA and mRNA profiling reveals potential regulatory network involved in SLE pathogenesis. (A) Heat map representation of differentially expressed genes in SLE patients and control subjects (n = 5). (B) Top 10 GO processes determined based on all differentially expressed genes highlight enrichment of neutrophil activation and chemokine activity. (C) Hierarchical clustering of 45 differentially expressed miRNAs in SLE patients and control subjects from the same cohort determined by mRNA sequencing (>2-fold change, P < 0.05). (D) The top four downregulated and upregulated miRNAs were analyzed by RT-qPCR in PBMCs from five SLE patients and five age- and gender-matched healthy controls. PBMC set different from that used in (C) was analyzed. Results are presented as the mean ± SEM. ***P < 0.001; **P < 0.01; *P < 0.05. (E) KEGG analysis of predicted target genes of miR-4512. (F) The regulatory networks of miR-4512 target genes and autoimmune disease-related genes.