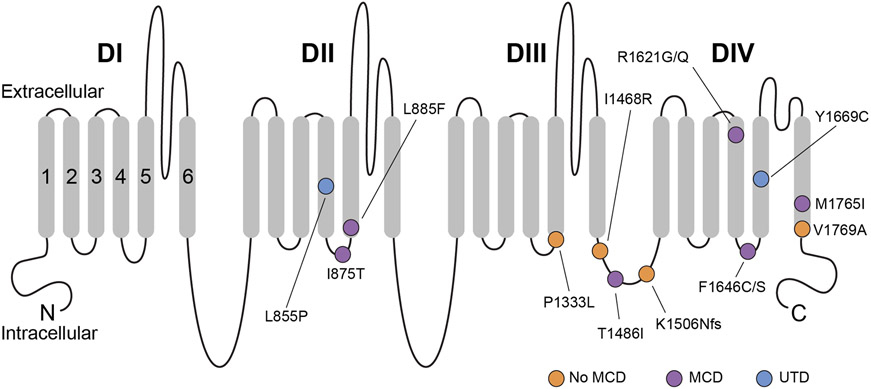
Figure 1. Schematic of the Nav1.3 protein.
Shown are all amino acid residues corresponding to disease-associated pathogenic or likely pathogenic variants in SCN3A. Nav1.3 is formed by four repeated domains (DI-IV), each composed of six transmembrane segments (S1-6), with S1-4 of each domain mediating voltage sensing and S5-6 forming the ion conducting pore. Variants associated with malformation of cortical development are indicated with a purple circle, while variants identified in patients without malformation of cortical development are indicated with an orange circle. A blue circle denotes variants identified in patients where neuroimaging data was unavailable. Note that pathogenic variants appear to be preferentially clustered in S4-6 of DII-IV, with many variants at/near the intracellular S4-5 linker of DII-IV. MCD, malformation of cortical development. UTD, unable to determine (due to absence of available neuroimaging data).