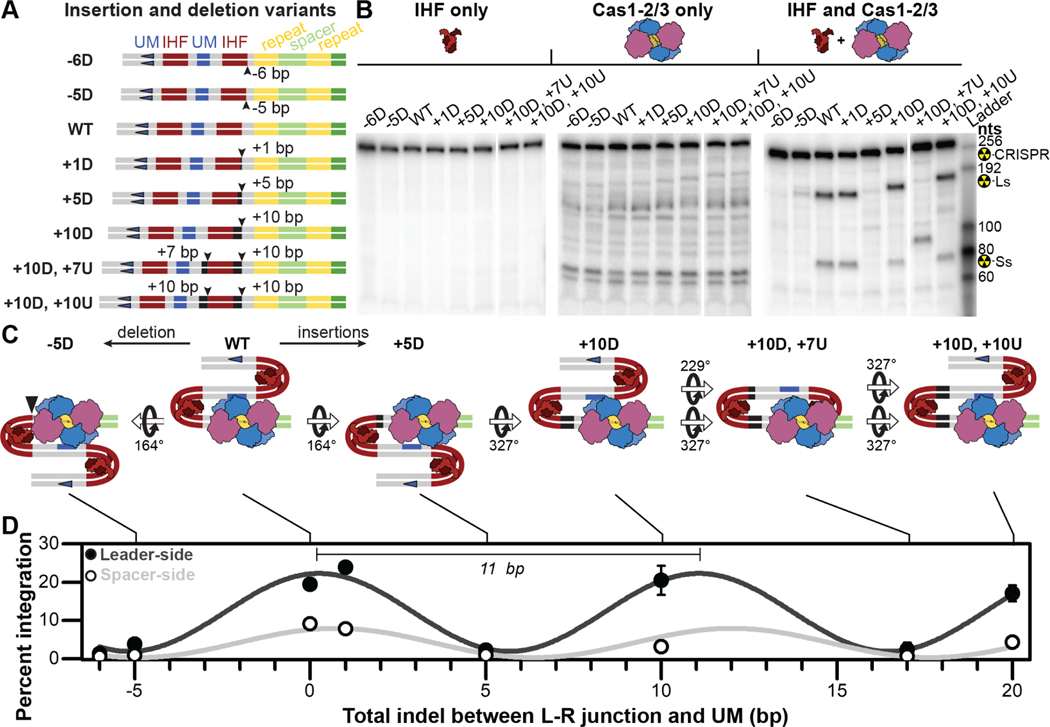
Figure 4. Phase-dependent CRISPR adaptation.
(A) Schematic of insertion and deletion (indel) variants generated to test the impact of distance and phase of leader motifs relative to the CRISPR locus. (B) I-F CRISPR integration is insensitive to indels that maintain phase, (10-base pairs), while indels of less than a full helical turn (five to seven base pairs) result in integration defects. Representative images of integration assays resolved on denaturing gels. Reactions were performed either in the presence of IHF alone (left), Cas1-2/3 alone (middle), or in the presence of both protein complexes (right). The expected positions of full-length substrate (249–275 nts depending on indel), as well as leader-side (145–161 nts depending on indel) and spacer-side (77 nts for all variants) integration products are indicated. (C) Schemes of putative IHF-bound conformations of wildtype and mutant leaders. Estimated rotations of DNA, relative to wildtype, are indicated. (D) Quantification of prespacer integration into all leader indel variants, either at leader- (dark gray) or spacer-side (light gray). The average (±s.d.) of triplicate reactions is shown. Leader- and spacer-side integration data were independently fit to sine waves (R2=0.95 and R2=0.75), of wavelengths 10.8 ± 0.4 and 11.3 ± 0.7 bp, respectively. Integration was confirmed via high-throughput sequencing (Figure S3). Uncropped images and replicate gels are provided (Figure S6). See also Figure S3 and S6.