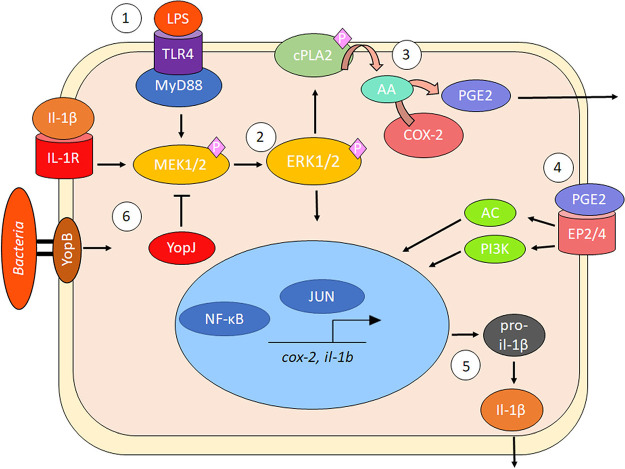
FIG 9.
Model of PGE2 and IL-1β biosynthesis in macrophages during Gram-negative bacterial infection. (Step 1) LPS ligand binding to TLR4 induces rapid phosphorylation of host MAPKs, such as MEK1/2, with adaptor protein assistance, including MyD88. (Step 2) MEK1/2 becomes activated and phosphorylates ERK1/2, leading to the activation and migration of transcription factors to the nucleus to induce proinflammatory gene transcription, including upregulation of COX-2 transcripts. (Step 3) Activated ERK1/2 phosphorylates and activates cPLA2 at Ser 515/505 residue, resulting in the liberation of AA from membrane phospholipids and AA’s conversion into PGH2/PGE2 by COX-2. (Step 4) PGE2 is secreted and binds in an autocrine fashion to specific EP2/4 receptors to activate IL-1β transcription by altering cAMP levels or PI3K-induced migration of transcription factors NF-κB and AP-1. In the presence of Gram-negative bacteria, pro-IL-1β is cleaved to mature IL-1β by caspase-1 due to inflammasome activation. (Step 5) IL-1β forms a positive-feedback loop with PGE2 by binding to IL-1R and increasing COX-2 transcription. Y. pseudotuberculosis and S. flexneri modulate inflammasome activation and PGE2 biosynthesis by manipulating host MAPK/ERK-dependent signaling such as T3SS factors YopJ (Y. pseudotuberculosis) or OspB/OspF (S. flexneri).