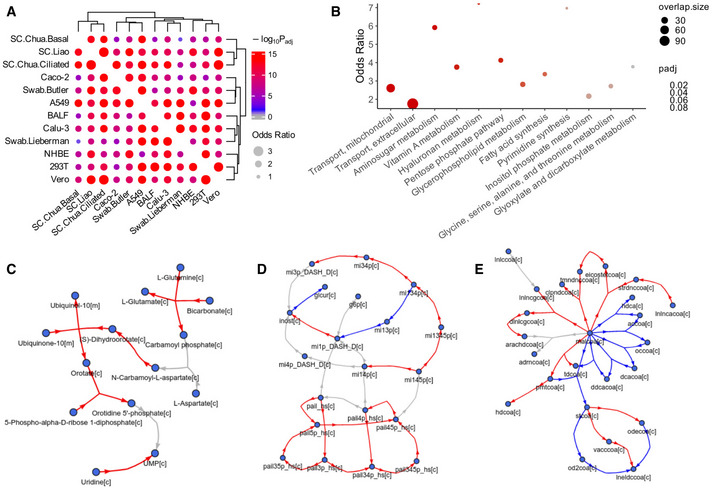
Visualization of the relatively consistent DF patterns in selected enriched pathways. The DF results are based on metabolic modeling using the human GEM Recon 3D (Brunk
et al,
2018), but for clear visualization, the metabolic network graphs are based on the human GEM Recon 1 (Duarte
et al,
2007) to reduce the number of metabolites and reactions displayed (
Materials and Methods). Metabolites are represented by nodes, reactions are represented by directed (hyper) edges, with edge direction corresponding to the consensus reaction direction and edge color corresponding to the consensus DF direction across datasets (
Materials and Methods). Red and blue colors correspond to increased and decreased fluxes, respectively; gray color corresponds to reactions not showing consistent DF changes across datasets, some of such reactions are not shown to increase clarity. (C) Pyrimidine synthesis. (D) Inositol phosphate metabolism. (E) Fatty acid synthesis. Metabolites are labeled by their names in (C) or IDs in (D, E), with suffixes denoting their cellular compartments: [c] cytosol; [m] mitochondria. The mapping between the IDs and metabolite names in (D, E) is given in Table
EV5C.