FIG 1.
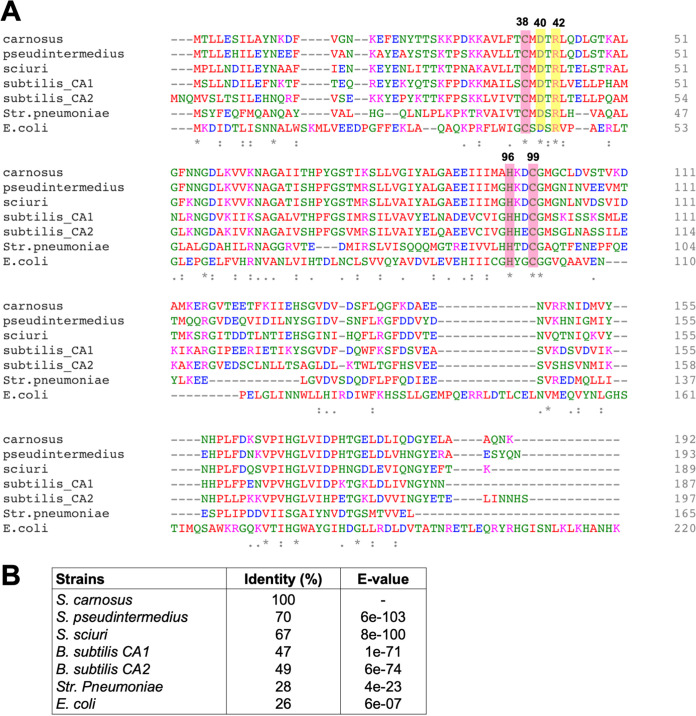
Multiple protein sequence alignment for CA. (A) The alignment was carried out for the CAs from S. carnosus TM300 (carnosus), S. pseudintermedius ED99 (pseudintermedius), S. sciuri FDAARGOS_285 encoded by locus tag Ga0225916_842 with Pfam00484 (sciuri_CA), Bacillus subtilis subtilis 168 encoded by locus tag BSU30690 with Pfam00484 (subtilis_CA1), B. subtilis 168 encoded by locus tag BSU34670 with Pfam00484 (subtilis_CA2), Str. pneumoniae TIGR4 encoded by locus tag SP0024 (pneumoniae), and E. coli MG1655 encoded by locus tag b0126 (E. coli). All sequences were obtained from the IMG/M database. The deduced sequences were aligned using Clustal Omega (56). The numbers correspond to the S. carnosus sequence. Highlighted in red are residues 38 (cysteine), 96 (histidine) and 99 (cysteine), which are ligands to zinc. Residues 40 and 42 highlighted in yellow are an Asp/Arg dyad that is important for the proton transfer step of catalysis. (B) The percentages of identity of the CA protein from S. carnosus to those of the other sequences were compared using BLASTp (51).