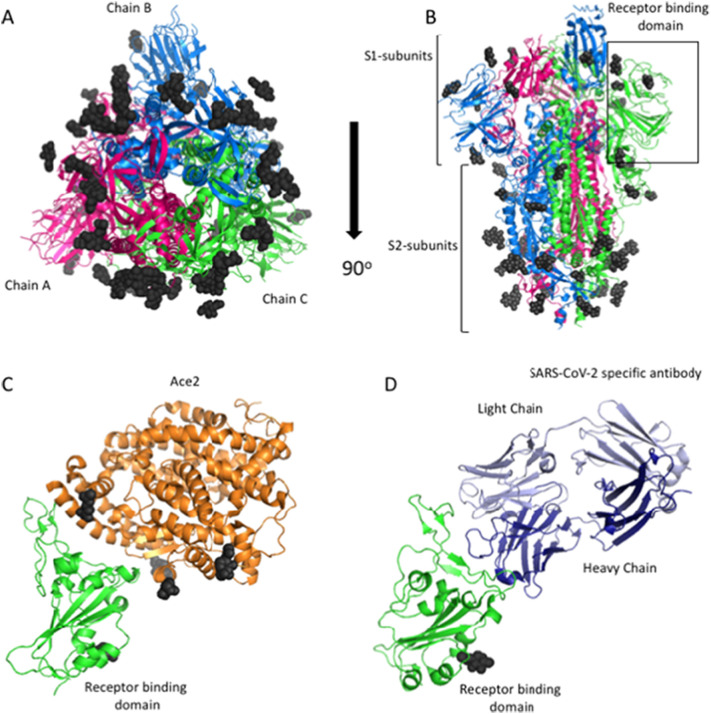
Fig. 7.
Structures of the SARS-CoV-2 S-glycoprotein. A) Cartoon diagram of the 3D structure of the S-glycoprotein solved using cryo-EM to 3.46 Å (PDB ID: 6VSB). The S-glycoprotein forms a trimer composed of an S1-subunit (pink), an S2-subunit B (green) and an S3-subunit (blue). This structure represents the prefusion confirmation and was found to have 22 glycosylation sites (dark grey). B) 90° rotation of A. Here, the receptor binding domain (RBD) is located at the top of the structure. The RBD of the subunit C is boxed in black. C) Cartoon diagram of the RBD of the S-glycoprotein (green) bound to the host receptor ACE2 (orange) (PDB ID: 6M0J). This structure was solved to 2.45 Å using X-ray protein crystallography. D) Cartoon diagram of the SARS-CoV-2 RBD (green) bound to a SARS-CoV-2 specific antibody (the antibody heavy and light chains are colored dark blue and light blue, respectively). This structure was solved using X-ray crystallography to 2.85 Å (PDB ID: 7BWJ). It is important to note that the antibody binding site is at the same position as the ACE2 binding site, thereby blocking interactions between the virus and the host receptor. All figures were made using PyMol. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)