FIG 4.
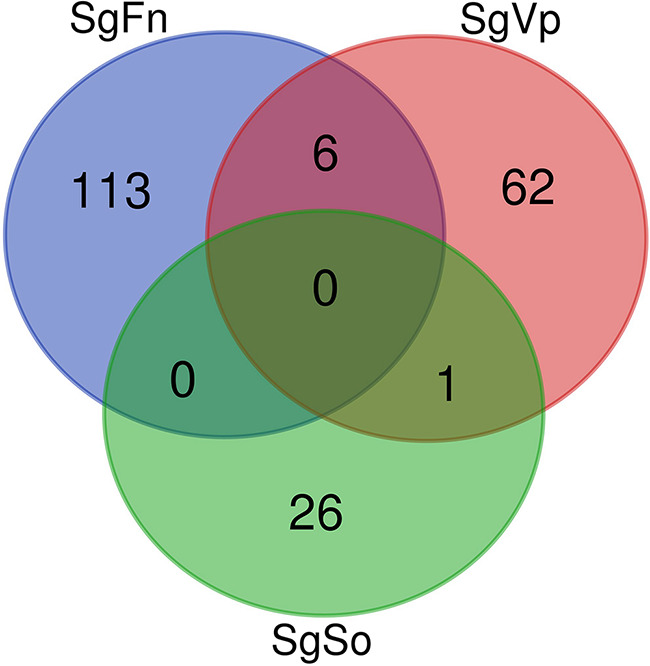
Venn diagram showing overlaps between three S. gordonii pairings. In total, six genes were common between S. gordonii-Fusobacterium nucleatum (SgFn) and S. gordonii-Veillonella parvula (SgVp), whereas only one gene was found in common between SgVp and S. gordonii-S. oralis (SgSo), and there were no genes in common between the SgSo versus SgFn pairings (see Table S2 in the supplemental material). Among the six genes that were regulated in SgFn and SgVp pairings, four genes were regulated in the same direction in two different bacterial pairings. These genes encode the tagatose-6-phosphate kinase (fold change = −3.2 in SgFn and −2.72 in SgVp), truncated hypothetical protein (fold change = 2.5 in SgFn and 2.62 in SgVp), short-chain dehydrogenase (fold change = −2.32 in SgFn and −2.75 in SgVp), and thiamine biosynthesis protein (fold change = −2.46 in SgFn and −2.04 in SgVp). Interestingly, two genes were regulated in the reverse direction in two different bacterial pairings. These genes encode the recombination regulator SGO_RS03085 (fold change = 2.84 in SgFn and −2.88 in SgVp) and Fur family transcriptional regulator (fold change = 2.42 in SgFn and −2.68 in SgVp). The one gene (pf08796 family protein) that was common between the SgSo and SgVp pairings was regulated in the same direction in both pairings.
