Figure 1. Construction of Tg (0.9 lbx1a:EGFP)SU32 and Tg(1.6 lbx1a:EGFP)SU33 transgenic lines.
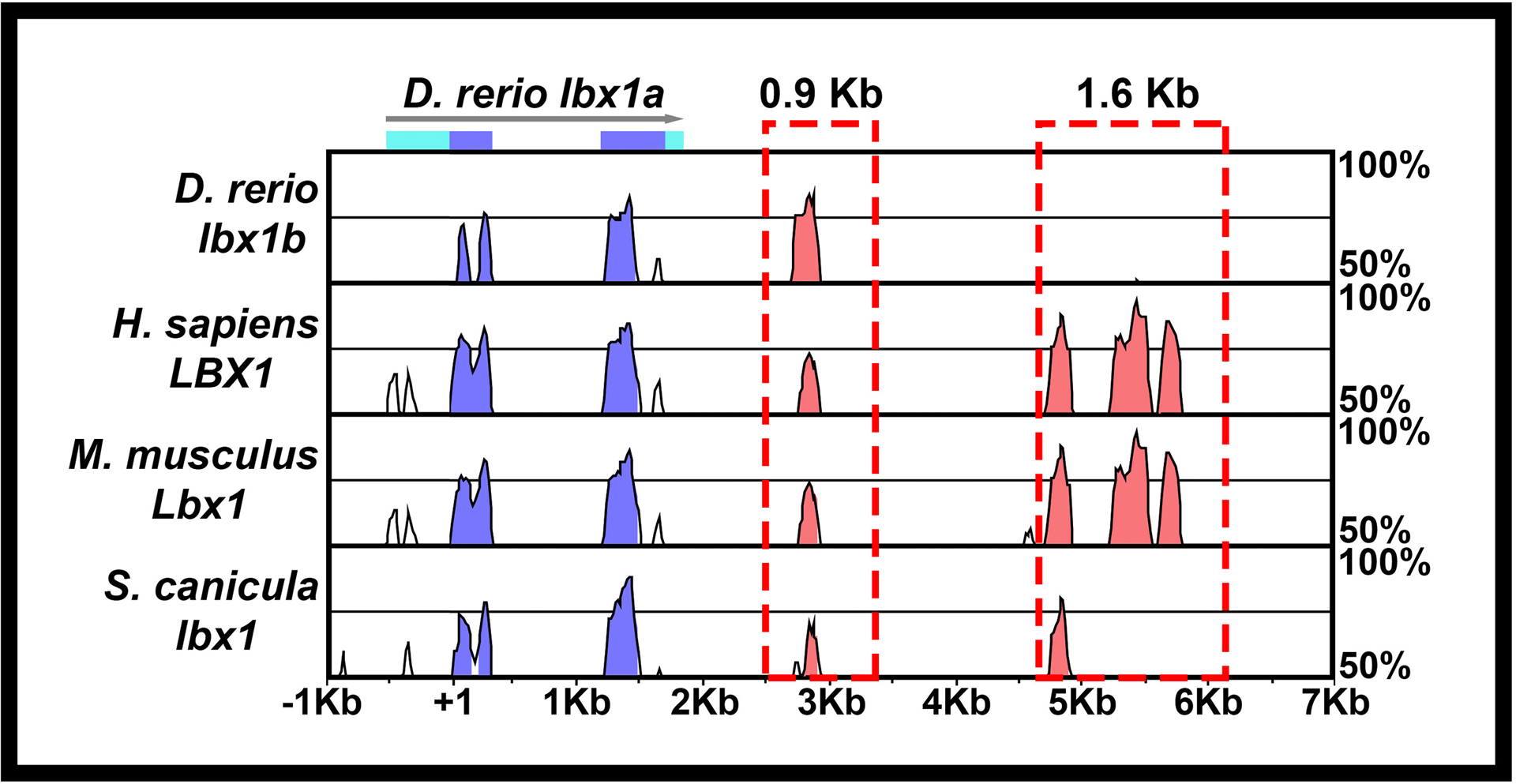
Schematic showing Shuffle-MLAGAN analysis of Danio rerio lbx1a genomic region with zebrafish sequence as baseline compared to Danio rerio lbx1b genomic sequence and orthologous regions in mouse and humanHomo sapiens, Mus musculus and Scyliorhinus canicula genomes. Conserved coding sequences are indicated in blue, arrow indicates 5’→3’ orientation, light blue boxes indicate untranslated regions of D. rerio lbx1a. Conserved non-coding elements (CNEs) in 3’ region are indicated in pink. The 0.9 Kb and 1.6 Kb regions amplified to create the Tg(0.9 lbx1a:EGFP)SU32 and Tg(1.6 lbx1a:EGFP)SU33 transgenic lines areis indicated with a red dotted boxes.
