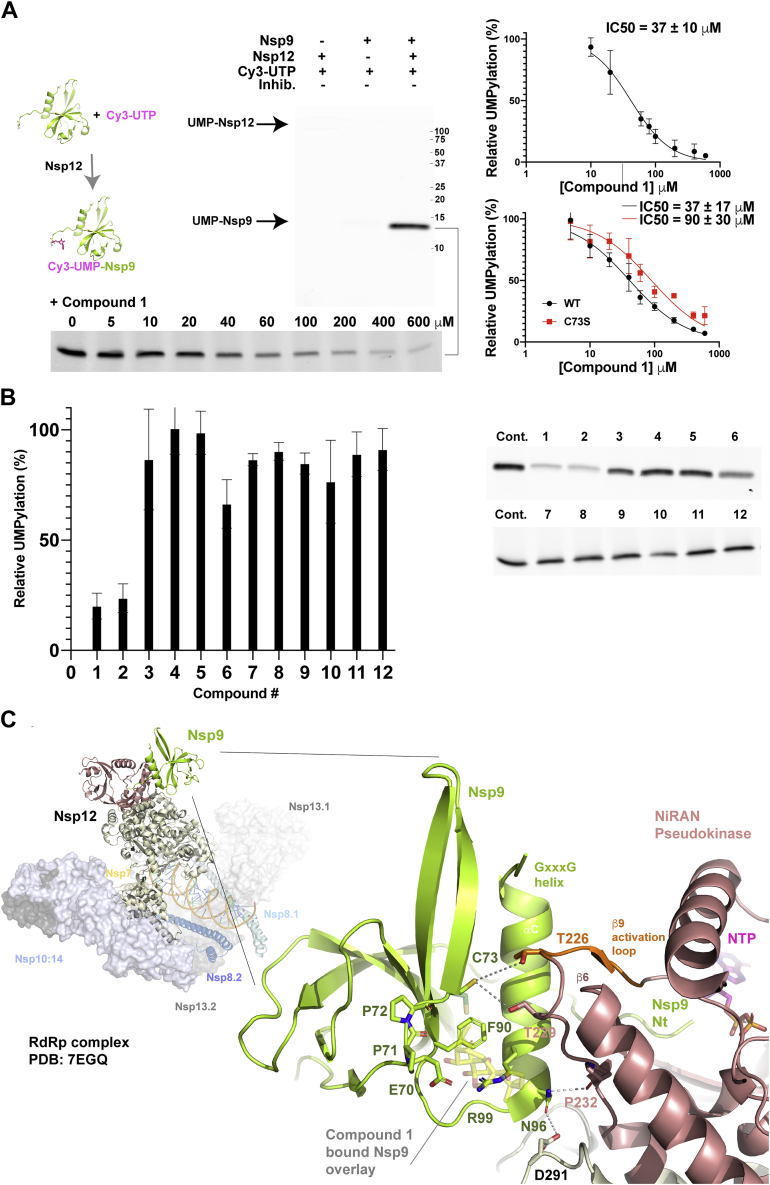
Figure 4.
Oridonin binding mapped onto the SARS-CoV-2 extended replication transcription complex.A, UMPylation assay of Nsp9 by the NiRAN domain of Nsp12. Cy3-labeled UTP was incubated with Nsp9 and Nsp12 with the reaction products, then separated by SDS-PAGE, and imaged. IC50 calculations were obtained from a control-normalized dose–response fit to the data. Errors represent the S.D. of triplicate reaction series. B, the UMPylation reaction was repeated in triplicate, the presence of each of the 12 compounds described previously and compared with in-gel control reactions. Quantification of the Cy3-modified Nsp9 is plotted with errors representing standard deviation of three independent reactions. Note: the upper gel in panel A and cropped compound screen 1 to 6 of panel B are part of the same experiment. C, a cartoon representation of the extended RTC (26) is shown with the Nsp9: NiRAN-domain interaction highlighted. Nsp12 is colored with salmon for the pseudokinase domain and sand for the RdRp. The pseudokinase activation loop is in orange. The engaged Nsp9 is colored green with N-terminal residues labeled. The oridonin-binding site is overlaid in partial transparency onto the RNA complex indicating potential clashes with residues immediately C-terminal to the activation loop and near the Asn-96 engagement point. Potential Nsp9:Nsp12 interface interactions are highlighted with gray lines, the local resolution of cryo-EM map (26) identifies their proximity but precludes specific categorization into, e.g., van der Waals contacts or hydrogen bonds.