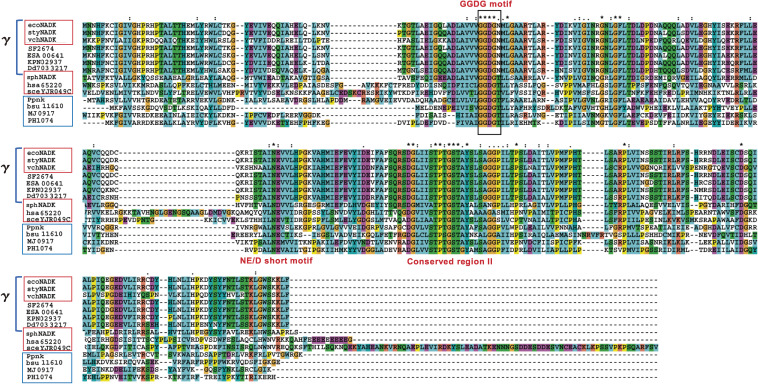
Figure 2.
Multiple alignment of the primary structure of ATP-NAD kinases (boxed in red), ATP/poly(P)-NAD kinases (boxed in blue), and γ-proteobacterial NAD kinase homologs (indicated by γ).62) ATP-NAD kinases are identified as follows: ecoNADK (E. coli), styNADK (S. enterica), vchNADK (V. cholerae), sphNADK (Sphingomonas sp. A1), hsa65220 (human), and sceYJR049C (S. cerevisiae). ATP/poly(P)-NAD kinases are Ppnk (M. tuberculosis), bsu11610 (B. subtilis), MJ0917 (M. jannaschii), and PH1074 (P. horikoshii). γ-Proteobacaterial NADK homologs are ecoNADK, styNADK, vchNADKs, SF2674 (Shigella flexneri), ESA00641 (Cronobacter sakazakii), KPN02937 (Klebsiella pneumoniae), and Dd703 3217 (Dickeya dadantii). The motifs GGDGN and GGDGT are boxed in black. The motif GGDGN is conserved in the primary structures of some γ-proteobacterial NAD kinase homologs, while the motif GGDGT is found in those of other ATP-NAD kinases and ATP/poly(P)-NAD kinases. Identical residues are denoted by an asterisk (*), strongly conserved residues by a colon (:), and weakly conserved residues by a period (.). This figure was reproduced from Y. Nakamichi et al. (2013) Sci. Rep. 3, 2632 (Ref. 62, Supporting information) with some modifications.