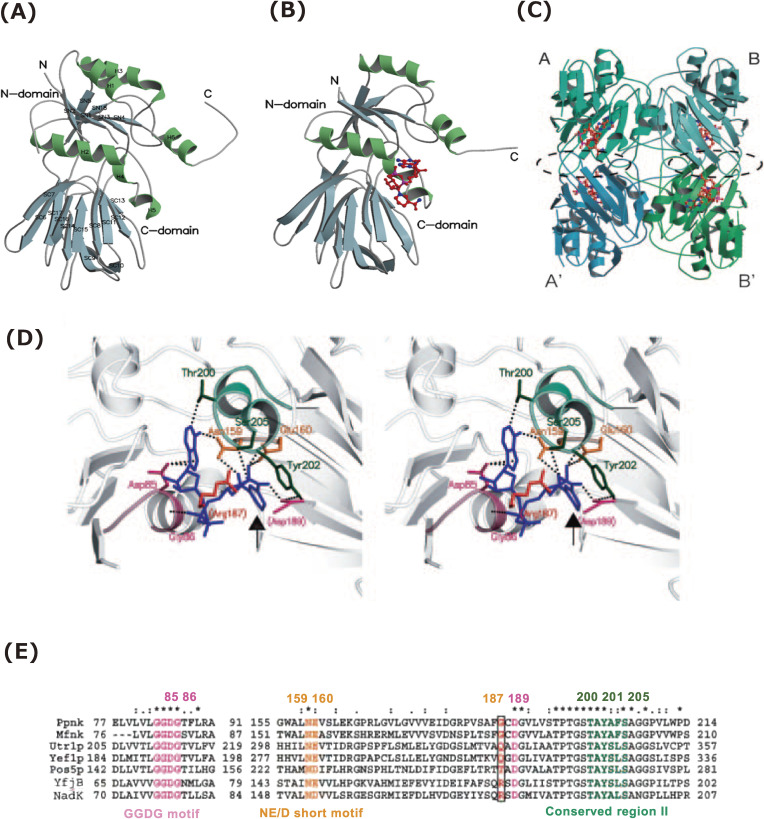
Figure 4.
Structure of NAD kinase and structural determinant of phosphoryl acceptor specificity.39–41,63) (A) A ribbon model of apo-Ppnk. The α-helix (H) and β-strand (SN and SC for N- and C-domains) are designated by numbers from N-terminal. (B) A ribbon model of holo-Ppnk (Ppnk-NAD). NAD is indicated in red (nitrogen, blue; phosphorus, pink). (C) A ribbon model of the quaternary structure of Ppnk-NAD. An asymmetric unit contains two subunits (Ppnk-NAD-A/-B or -A′/-B′). NAD+ is shown to be similar to (B). The A-A′ and B-B′ intersubunit contact regions are enclosed by a broken black line. (D) Stereodiagram of the binding site for NAD+ (blue) in Ppnk with the exception of Arg-187. Arg-187 is artificially replaced with Gly-187 in Ppnk. Asp-189 and Arg-187 are provided by the adjacent subunit and are denoted in parentheses. Residues of Gly-83 to Gly-86 (GGDG motif), Asn-159 to Glu-160 (NE/D short motif), and Thr-200 to Ser-205 (in conserved region II) are indicated in purple, yellow, and green, respectively. Contacts are represented by dotted lines. The nicotinamide ring of NAD+ is emphasized using an arrow. (E) Multiple alignment of the primary structures of NAD(H) kinases (Ppnk, Mfnk, Utr1p, Yef1p, and Pos5p) and NAD kinases [YfjB (ecoNADK) and NadK (sphNADK)]. The primary structures corresponding to the NAD+-binding site of Ppnk are aligned using ClustalW. The number of residues for each enzyme is specified. The residues participating in NAD+ binding are colored as in (C), while the important residues corresponding to Gly-187 in Ppnk are boxed. Identical residues are denoted by an asterisk (*), strongly conserved residues by a colon (:), and weakly conserved residues by a period (.). This figure was reproduced from S. Kawai and K. Murata (2008) Biosci. Biotechnol. Biochem. 72, 919–930 (Ref. 63) with some modifications.