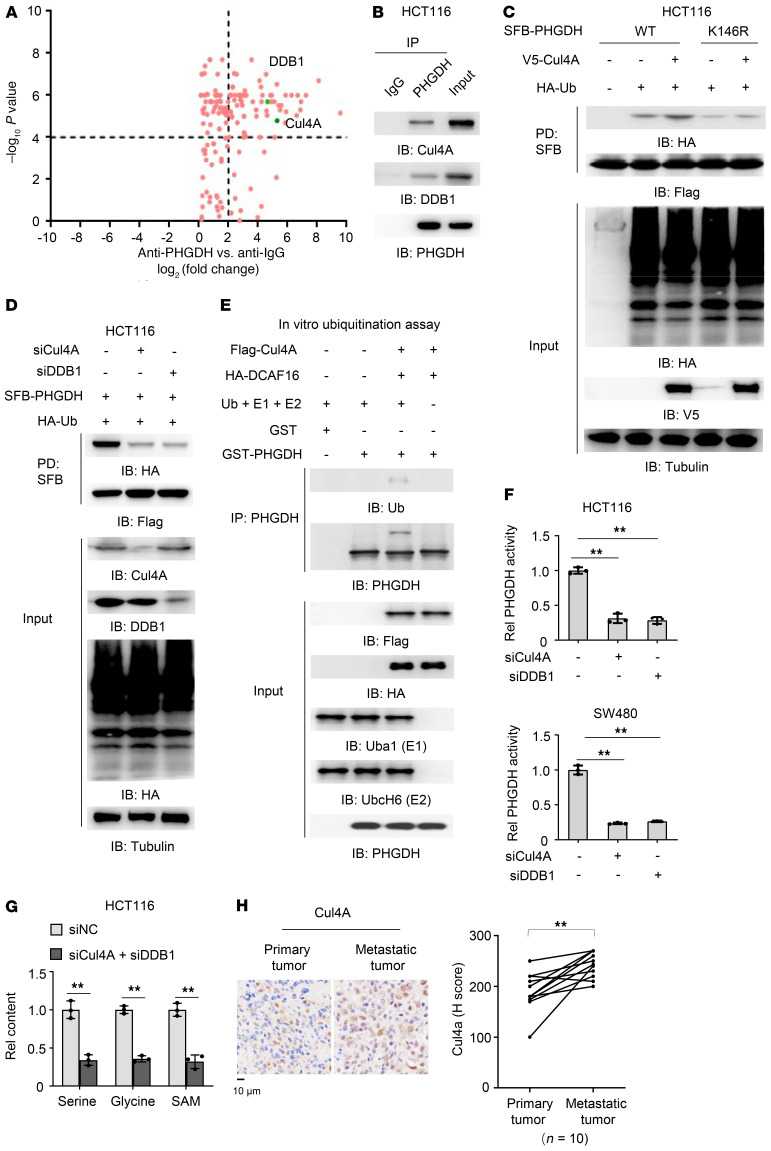
Figure 3. Cul4A–DDB1–CUL4-associated factor 16 complex mediates the monoubiquitination of PHGDH.
(A) PHGDH proteins were immunoprecipitated from HCT116 cells and the immunoprecipitated complex was further analyzed by liquid chromatography–tandem mass spectrometry for identification of PHGDH-associated proteins. Data represent the means ± SD of 2 independent experiments. (B) HCT116 cells were harvested and PHGDH proteins were immunoprecipitated. (C) HCT116 cells stably expressing SFB-PHGDH WT or K146R were cotransfected with EV or HA-Ub and EV or V5-Cul4A. SFB-PHGDH proteins were pulled down using streptavidin agarose beads. (D) HCT116 cells stably expressing SFB-PHGDH were cotransfected with HA-Ub and siNC, siCul4A, or siDDB1. SFB-PHGDH proteins were pulled down using streptavidin agarose beads. (E) Flag-Cul4A and HA-DCAF16 were immunoprecipitated and purified from HEK293T cells stably expressing Flag-Cul4A or HA-DCAF16. In vitro ubiquitination assay was performed by incubating Flag-Cul4A and HA-DCAF16 with recombinant GST-PHGDH, His-ubiquitin (Ub), His-Uba1 (E1), and His-UbcH6 (E2). (F) HCT116 or SW480 cells stably expressing SFB-PHGDH were transfected with siNC, siCul4A, or siDDB1. SFB-PHGDH proteins were pulled down using streptavidin agarose beads for PHGDH activity measurement. One-way ANOVA with Tukey’s multiple-comparison test. (G) HCT116 cells were transfected with siNC or siCul4A and siDDB1. Cells were harvested for the measurement of intracellular glycine, serine, and SAM (2-tailed Student’s t test). (F and G) Data represent the mean ± SD of 3 biologically independent experiments. (H) IHC staining was performed in paired primary tumors and hepatic metastatic tumors from 10 patients with CRC with anti-Cul4A antibody. Staining scores of Cul4A were compared between primary tumors and metastatic tumors. Left, representative images of IHC staining were shown. Right, semiquantitative scoring was carried out (paired t test, 2 tailed). **P < 0.01. See also Supplemental Figure 3.