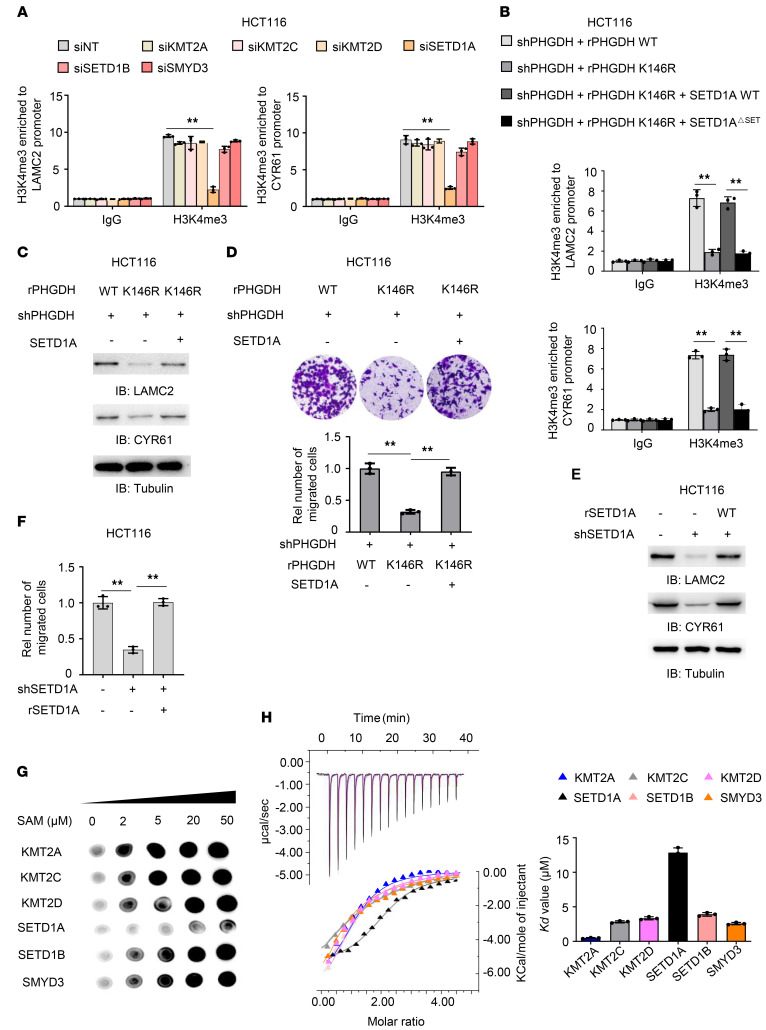
Figure 6. Increased SAM levels by PHGDH K146mUb promote tumor cell migration by initiating SETD1A-mediated histone methylation of LAMC2 and CYR61.
(A) HCT116 cells were transfected with siRNAs targeting nontargeting (NT), KMT2A, KMT2C, KMT2D, SETD1A, SETD1B, or SMYD3. ChIP analyses with anti-H3K4me3 antibody were performed. (B) PHGDH-depleted HCT116 cells rescued with rPHGDH WT or K146R were infected with or without the lentivirus expressing SETD1A or SETD1AΔSET. ChIP analyses were performed with anti-H3K4me3 antibody. (C and D) PHGDH-depleted HCT116 cells rescued with rPHGDH WT or K146R were infected with or without the lentivirus expressing SETD1A. Immunoblotting analyses were performed (C). Transwell migration assays were performed (D). (E and F) HCT116 cells were depleted of endogenous SETD1A and rescued with rSETD1A. LAMC2 and CYR61 expression was examined by immunoblotting analyses (E). Transwell migration assay was performed in these cells (F). (G) In vitro methylation assays were performed with purified recombinant His-tagged methyltransferase domains of KMT2A, KMT2C, KMT2D, SETD1A, SETD1B or SMYD3, SAM, and H3K4 peptide, followed by dot blot analysis with anti-H3K4me3 antibody. (H) ITC assays were performed with purified recombinant His-tagged methyltransferase domains of indicated HMTs and SAM. Representative images (left) and statistical results (right) of ITC were shown. (A, B, D, F, and H) Data represent the mean ± SD of 3 biologically independent experiments. One-way ANOVA with Tukey’s multiple-comparison test. **P < 0.01. See also Supplemental Figure 6.