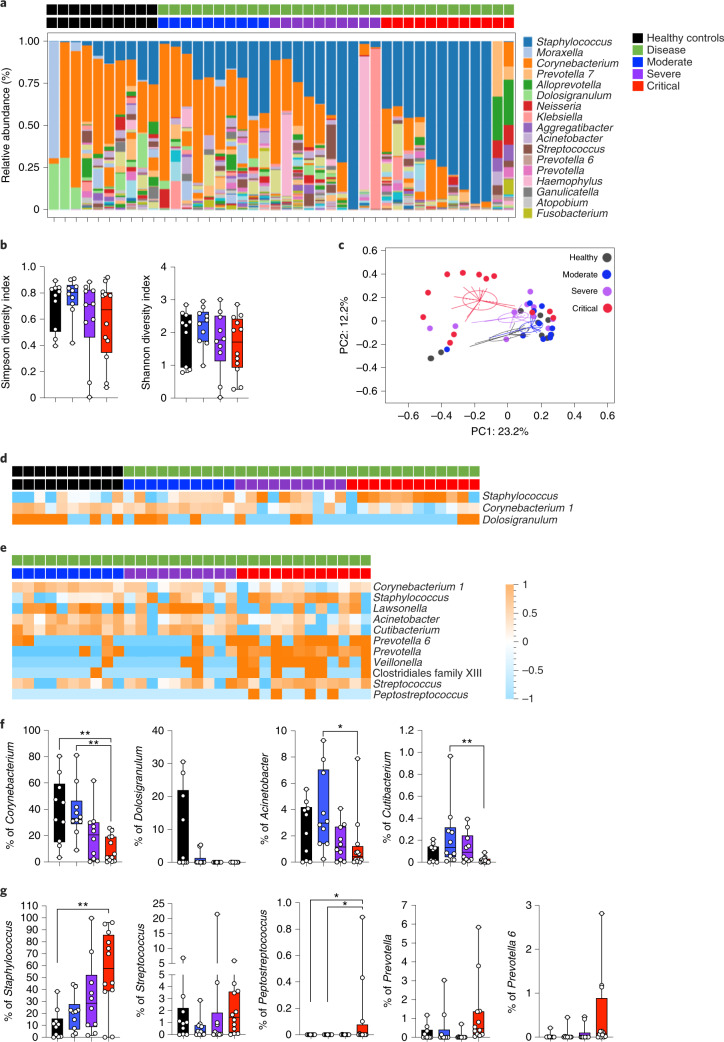
Fig. 5. Perturbations of nasopharyngeal 16S rRNA profiles in patients with COVID-19.
a–g, Nasopharyngeal bacterial communities were measured in healthy controls (n = 10) and in patients with mild-to-moderate (n = 10), severe (n = 10) and critical (n = 12) COVID-19. a, The percentage of relative abundance at the genus level. b, Shannon and Simpson diversity indices by patient severity. Data are presented as box plots with median ± minimum to maximum. c, Principal-component analysis of 16S bacterial profiles. d, Heat map of statistically different (P < 0.05) genus abundance between healthy controls and patients with COVID-19 (moderate, severe and critical). e, Heat map of statistically different (P < 0.05) genus abundance between patients with COVID-19 depending on disease severity. P values were determined with a two-tailed Mann–Whitney test. f,g, Plots showing the percentage of individual genus abundance by disease severity. In b, f and g, box-and-whisker plots show the minimum and maximum values, interquartile range and the median. Corynebacterium (critical versus healthy, P = 6.9 × 10−3; critical versus moderate, P = 2.6 × 10−3), Acinetobacter (critical versus moderate, P = 3.2 × 10−2), Cutibacterium (critical versus moderate, P = 6.6 × 10−3), Staphylococcus (critical versus healthy, P = 9.0 × 10−3), Peptostreptococcus (critical versus healthy, P = 3.3 × 10−2; critical versus moderate, P = 3.3 × 10−2). P values were determined with the one-sided Kruskal–Wallis test followed by Dunn’s post hoc test for multiple comparisons with Geisser–Greenhouse correction; *P < 0.05; **P < 0.01; ***P < 0.001. In e, z-score scale is indicated, with upregulation shown in orange and downregulation shown in blue.