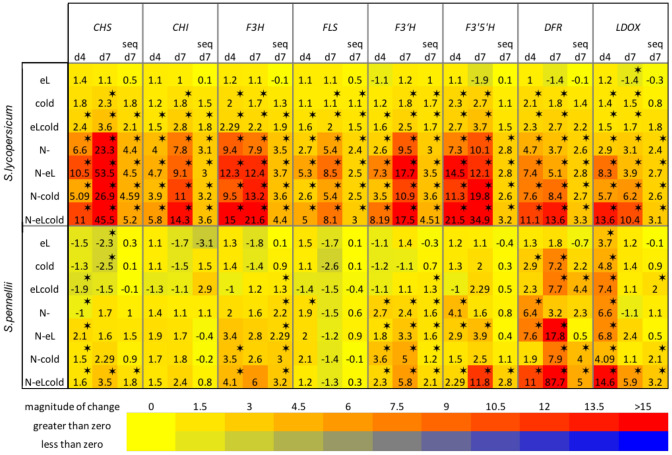
Fig. 6.
Expression of structural genes involved in secondary metabolism. RNASeq data were verified using samples from an independent biological experiment performed within the same plant chambers under the same stress conditions as before. S. lycopersicum and S. pennellii were grown on rockwool for 6 weeks before applying chilling temperatures (cold), elevated light intensities (eL), nitrogen deficiency (N-) or combinations thereof. 4 and 7 days after treatment induction the expression of indicated genes was determined in four biological replicates per treatment. Results are expressed as (fold change compared to the control sample). Significance is indicated for p < 0.001 as asteriks, or an FDR < 0.01 for RNASeq data. CHS chalcone synthetase, CHI chalcone isomerase, F3H flavanone-3-hydroxylase, FLS flavonol synthase, flavonoid-3-hydroxylase, flavonoid--hydroxylase, DFR dihydroflavonol-4-reductase, LDOX leucoanthocyanidin dioxygenase