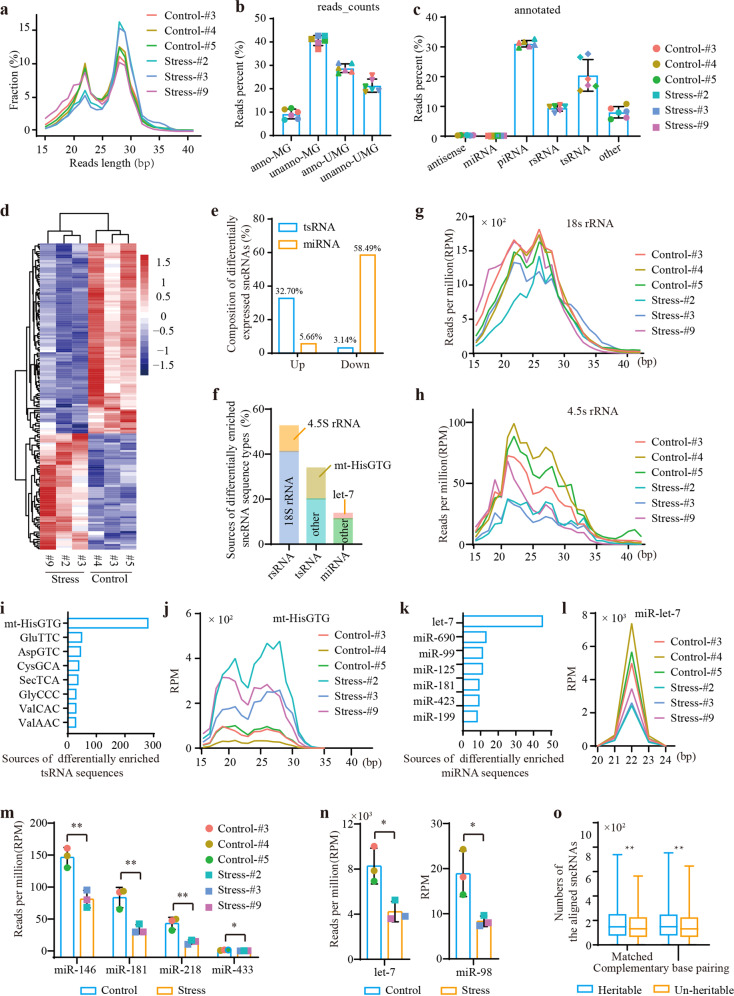
Fig. 6. Differentially enriched sncRNAs in paternal sperm.
a Length distribution of all sequencing reads. b Alignment of the sequencing data. anno_MG: matched to the reference genome and annotated; unanno-MG: matched to the reference genome but not annotated; anno-UMG: unmatched to the reference genome but annotated; unanno-UMG: unmatched to the reference genome and not annotated. c Distribution of the reads in annotated RNAs. d Heatmap of differentially enriched sncRNA subclasses. e Composition of the differentially enriched sncRNA subclasses. f Sources of differentially enriched sncRNA sequence types. g Length distribution of the 18S rRNA-derived rsRNA sequences in paternal sperm. h Length distribution of the 4.5S rRNA-derived rsRNA sequences in paternal sperm. i The main sources of the differentially enriched tsRNA sequences. j Length distribution of the mt (mitochondrial)-HisGTG-derived sequences. k The main sources of the differentially enriched miRNA sequences. l Length distribution of the miR-let-7-derived sequences. m Expression levels of the miRNAs related to Rhobtb3 gene. n Expression levels of the miRNAs related to Oprm1 gene. o Differences in the numbers of the aligned sncRNA sequences between heritable and un-heritable DMRs.