Figure 1.
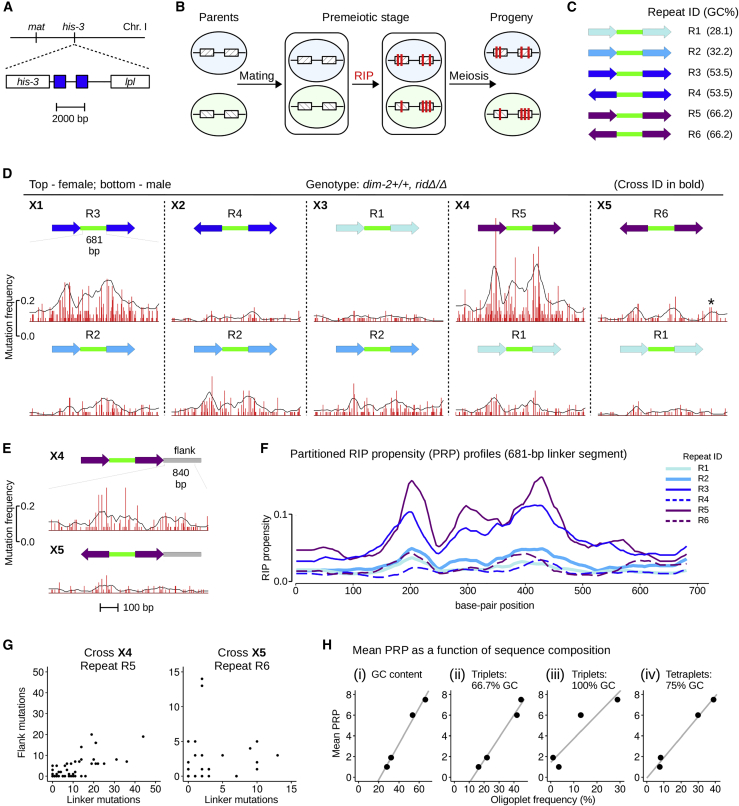
DIM-2-dependent RIP is affected by the GC content and the relative orientation of closely positioned perfect repeat units. (A) A pair of closely positioned perfect repeat units is integrated between his-3 and lpl genes on chromosome 1. The same integration site is used to create all repeat-carrying strains in this study (Table 2). (B) Mutation of two constructs can be assayed simultaneously in one cross. (C) Analyzed perfect repeats R1–R6 have the same overall length and the same linker sequence (green). Repeats differ with respect to GC content and relative orientation (Table 1). (D) Mutation of R1–R6 by DIM-2-dependent RIP was analyzed in five crosses (Table 3). Cross and repeat identifiers are indicated. 50 ascospore clones (per repeat construct per cross) were analyzed for RIP in the linker region. A 681-bp segment of the 729-bp linker was sequenced. The level of RIP is expressed as the number of mutations per site (“mutation frequency,” red) and as “partitioned RIP propensity” (PRP) profile (black). Data in (D and E) are plotted at the same scale (both x axis and y axis). (E) DIM-2-dependent RIP in the lpl-proximal flank was analyzed for the two GC-rich repeats R5 and R6 (crosses X4 and X5, respectively). (F) DIM-2-dependent PRP profiles for the linker region. Repeat identifiers are indicated. PRP profiles for repeats R1 and R2 are based on the combined data (crosses X3–X5 and X1–X3, respectively). (G) The relationship between the number of mutations in the linker versus the flank in individual spore clones for the GC-rich repeats R4 and R5. (H) Mean PRP as a function of sequence composition expressed as the overall GC content (i), the frequency of two triplet types (ii and iii) and the frequency of one tetraplet type (iv). To see this figure in color, go online.