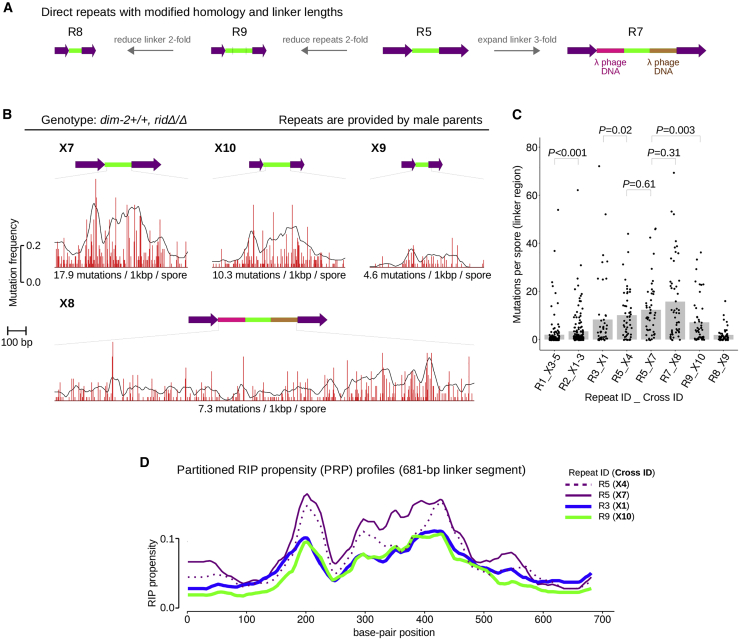
Figure 3.
DIM-2-dependent RIP of closely positioned repeats is modulated by the relative lengths of constituent segments. (A) Three additional repeats R7, R8, and R9 were constructed based on the GC-rich direct repeat R5. (B) Repeats were always provided by male parents (Table 3). 50 random ascospore clones were analyzed (per repeat per cross). The magnitude of RIP is expressed as in Fig. 1D. Data are plotted at the same scale as in Fig. 1D (both x axis and y axis). (C) Per spore numbers of DIM-2-dependent mutations in the sequenced linker segment. For repeats R1 and R2, the combined data sets are used. The mean number of RIP is indicated by a gray bar. The difference between empirical distributions of mutation counts is evaluated for significance by the Kolmogorov-Smirnov test. (D) DIM-2-dependent PRP profiles for the linker region. Repeat identifiers are indicated. To see this figure in color, go online.