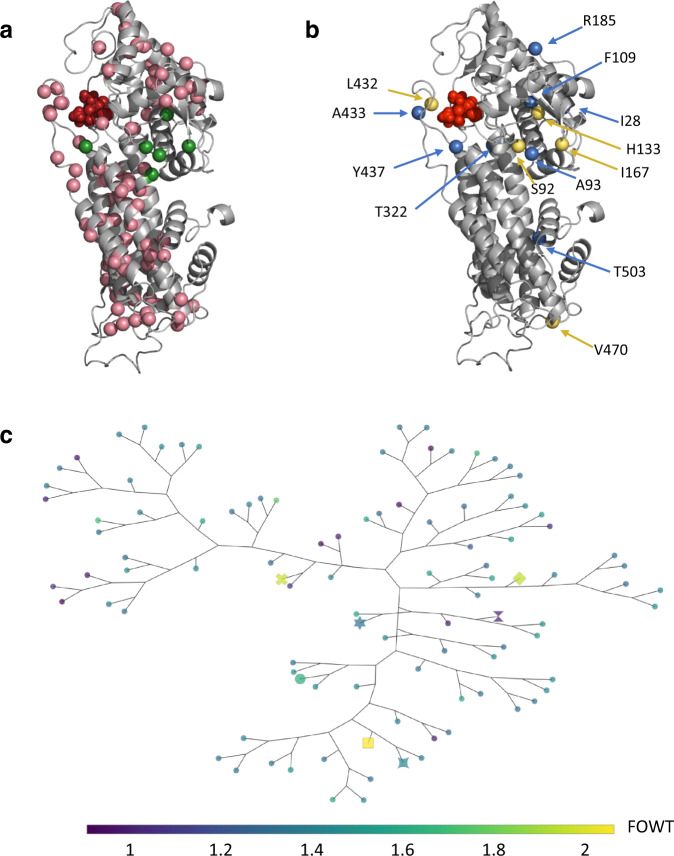
Fig. 3. Designed and enriched mutation positions.
Positions of interest are highlighted on a homology model generated for StlA using RosettaCM. Active site residues are highlighted in red. a Positions targeted for combinatorial mutagenesis during library construction. Any positions mutated in the library templates (see Methods) are highlighted in green, and additional mutations targeted during combinatorial mutagenesis are shown in pink. b Positions mutated in top hits. Positions mutated in the final SYNB1934 PAL variant are highlighted in yellow, and those mutated in other top hits (indicated with special markers in c, further detail in Supplement) are shown in blue. c A phylogenetic tree was generated by computing the distance between all the sequences using the Blosum62 substitution matrix57 and constructing the tree by neighbor-joining in Biopython58,59. Colors indicate activity level normalized to wild-type (FOWT). Markers: square = S92G_H133F_A433S_V470A, diamond = S92G_H133M_I167K_L432I_V470A, x = A93C_H133F_T322W_Y437N, star-square = I28R_S92G_H133F_V470A, large circle = S92G_H133F_R185E, hexagram = S92G_F109A_H133M_T503E, wild-type = hourglass; all other unique variants are shown as a small circle.