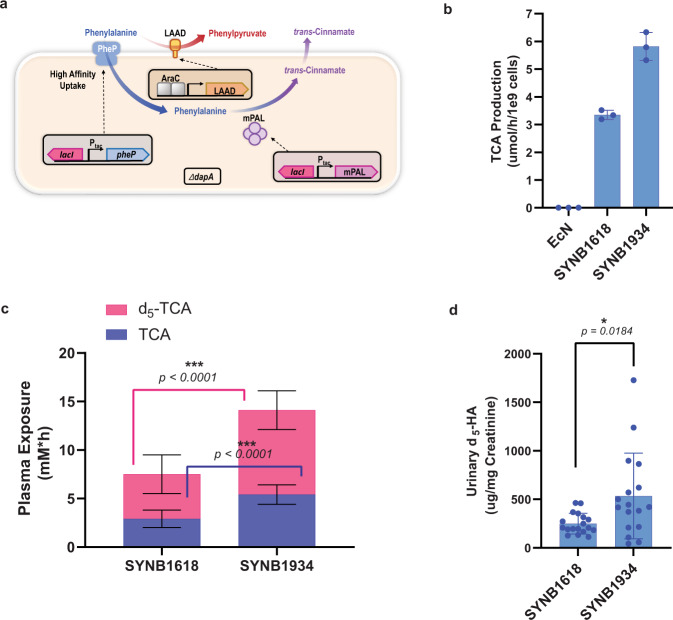
Fig. 5. In vitro and in vivo activity of SYNB1934.
Optimized PKU strain activity is compared to SYNB1618. a Schematic representation of the SYNB1934 bacterium shows key engineered elements, including the genes encoding mPAL, PheP, LAAD, and deletion of the dapA gene. b TCA production of 2.5 × 109 resuspended lyophilized SYNB1618 vs SYNB1934 cells was measured in an in vitro simulation of the gut environment. n = 3 independent biological triplicates ± s.d. c and d NHP subjects were dosed orally with a 5 g peptide and 0.25 g d5-Phe bolus followed by dosing with 1 × 1011 resuspended lyophilized SYNB1618 or SYNB1934 cells. Plasma areas under the curve (AUCs) for strain-specific biomarkers TCA and d5-TCA are displayed. For c, n = 18 biologically independent NHP subjects per group ± s.e. For each comparison, data were analyzed using a two-tailed unpaired t test (p < 0.0001). For urinary d5-HA concentration normalized to creatinine (d) n = 18 for SYNB1618-treated and 17 for SYNB1934-treated subjects ± s.e (one NHP in the SYNB1934-treated group failed to produce a urine sample over the course of experimentation). Data were analyzed using a two-tailed unpaired t test with Welch’s correction (p = 0.0184).