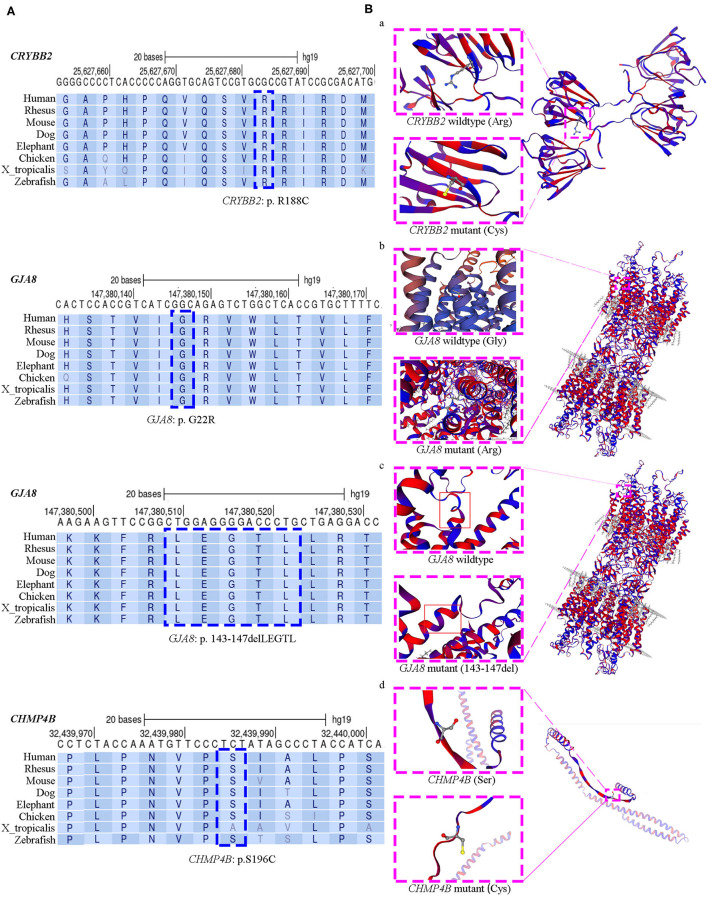
Figure 2.
Evolutionary sequence conservation analysis and structure homology modeling of mutant proteins and native proteins. (A) Multiplex sequence alignment of the βB2-crystalline protein, connexin 50, and the chromatin-modifying protein 4b from different species reveals that the four mutation (CRYBB2: R188C, GJA8: G22R, GJA8: p.143_147del LEGTL, and CHMP4B: S196C) are located within highly conserved regions (blue dashed box). (B) Structure homology modeling and comparison of mutant protein and native βB2-crystalline protein, Cx50, and the chromatin-modifying protein 4b (pink dashed box). The secondary structure was shown with α-helixes, β-sheets and loops in hydrophobic color scheme. From purple to red shows the least hydrophobic residues to most hydrophobic residues. (a) In the missense CRYBB2: p.R188C, the substitution replaces a basic arginine residue with an uncharged cysteine residue at the surface of the βB2-crystalline protein; (b) In the missense GJA8: p.G22R, a buried uncharged glycine residue mutants to a charged arginine residue, which located at the first transmembrane region of Cx50; (c) The small deletion GJA8: p.143_147delLEGTL locates at the cytoplasmic loop of the Cx50 protein, close to the third transmembrane domain; (d) The CHMP4B: p.S196C mutation is a substitution of serine residue to cysteine residue, which is located in a random coil region. This mutation makes a new exposed sulfhydryl group.