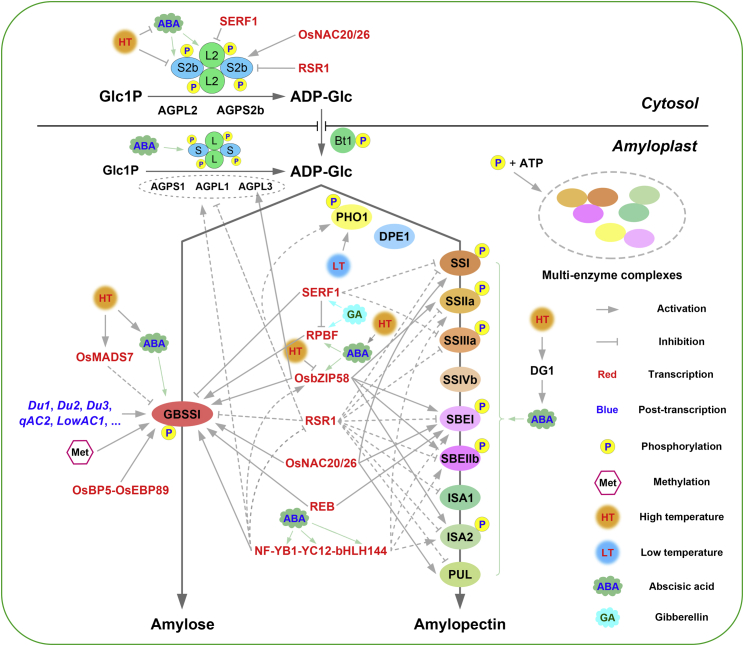
Figure 3.
Regulatory network of starch synthesis in the cereal endosperm: rice as an example.
At the transcriptional level, several key transcription factors (red font) and methylation are involved in the stimulation or inhibition of starch synthesis-related genes (SSRGs). At the post-transcriptional level, some Du (Dull) genes (Du1, Du2, Du3, etc.) and quantitative trait loci (QTLs) (qAC2, LowAC1, etc.) have been reported to regulate amylose synthesis by manipulating the splicing efficiency of Wx mRNA. At the post-translational level, phosphorylation affects starch synthesis by promoting the formation of multi-enzyme complexes. Gibberellin (GA) modulates the expression of SERF1 and RPBF in starch synthesis to promote starch accumulation in the developing rice endosperm. Abscisic acid (ABA) synthesized in rice and maize leaves directly activates the expression of most SSRGs and multiple hub transcription factors in the rice caryopsis after long-distance leaf-to-caryopsis ABA transport. Defective grain-filling 1 (DG1)-mediated long-distance ABA transport is sensitive to high temperature (HT) during seed development. HT also downregulates the expression of SSRGs and the stability of their encoded enzymes, especially genes encoding AGPase and GBSSI, resulting in chalky grains and defective starch accumulation. Low temperature (LT) promotes the function of PHO1. The arrowed lines indicate activation, and the bar-ended lines indicate inhibition. The dotted lines indicate that the transcription factor can regulate the transcription of the target gene, but there is no direct interaction evidence. The abbreviations for the enzymes, transcription factors, and genes are the same as those listed in Tables 1 and 2.