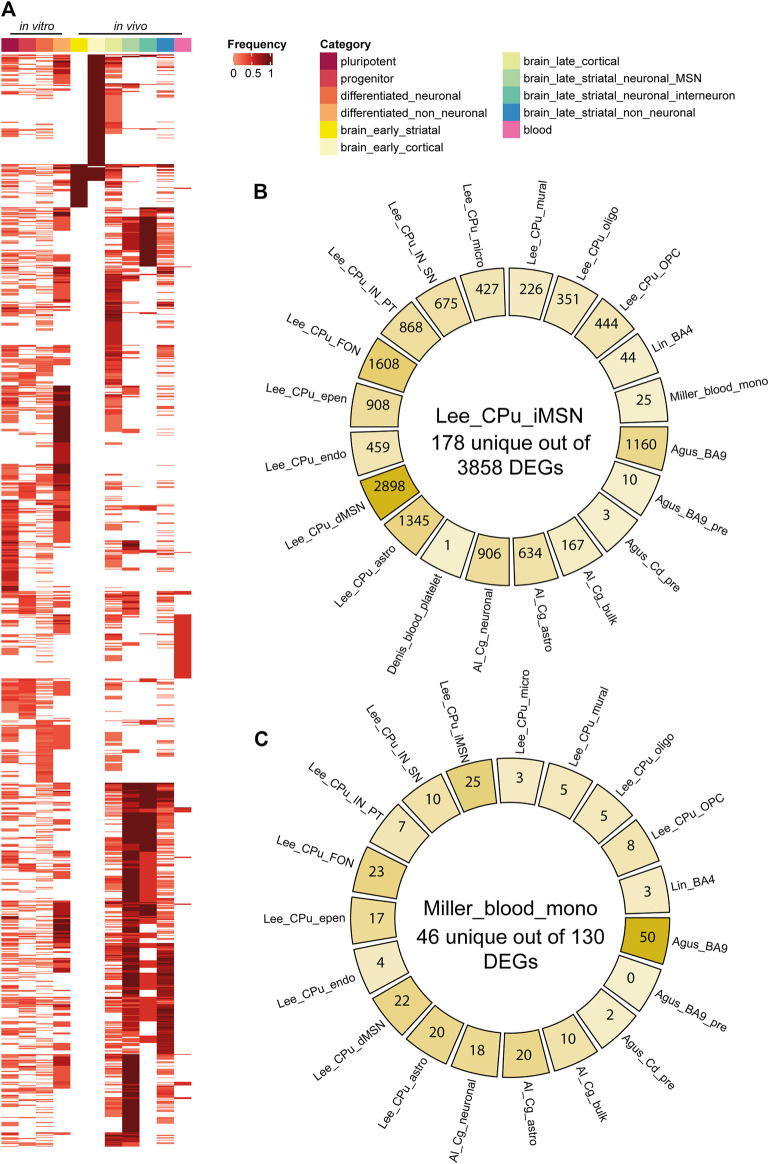
FIGURE 3.
HD differentially expressed gene (DEG) comparisons between categories and datasets. (A) DEG Frequency heatmap across all subcategories. Each column is a subcategory, each row is a DEG. DEG frequency is calculated as the number of datasets that DEG appears in divided by the total number of datasets in that subcategory. (B,C) Diagrams for pairwise comparisons of indirect pathway medium spiny neuron (iMSN) or blood monocyte DEG list vs all other data from primary tissues/cells. At the center is the number of unique DEGs in iMSN or blood monocyte, and on the side is the number of DEGs in common with other datasets. Abbreviations: CPu, caudate putamen; astro, astrocyte; epen, ependymal; endo, endothelial; IN, interneuron; PT, Pvalb/Th-expressing; MSN, medium spiny neuron; MNS, MAP2+ and NES/SOX2-; OPC, oligodendrocyte progenitor cell; oligo, oligodendrocyte; micro, microglia; NSC, neural stem cell; Cg, cingulate cortex; FON, Foxp2/Olfm3-expressing neuron; SN, Sst/Npy-expressing; dSPN/iSPN, direct/indirect pathway spiny projection neuron; mono, monocyte; NPC, neural progenitor cell; CoNeuron, cortical neuron; Cd, caudate nucleus; pre, presymptomatic; GPC, glial progenitor cell; APC, astrocyte precursor cell; BBB, blood brain barrier.