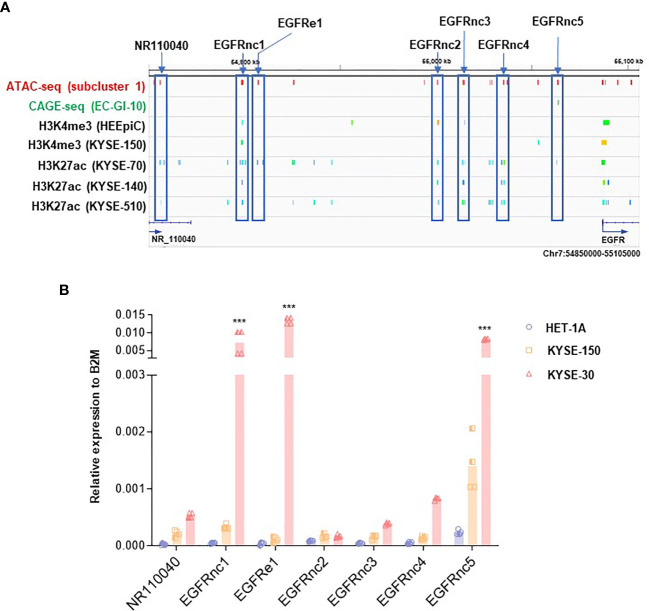
Figure 3.
Identification of non-coding RNAs (ncRNAs) near EGFR upregulated in KYSE-30 cells. (A) Multi-omics analysis approach revealed non-coding RNAs upstream of EGFR that are overlapped with both hyper-accessible regions in KYSE-30 cells (ATAC-seq peaks) and potential enhancer regions in other ESCC cells (CAGE-seq or H3K27ac peaks). Various colors in each ChIP-seq row indicate different peaks that are generated from one sequencing result. Blue boxes highlight the non-coding RNAs tested in this study. (B) Expressions of ncRNAs in HET-1A, KYSE-150, and KYSE-30 cells were measured by digital droplet PCR, and their copy numbers relative to the copy numbers of B2M, a commonly used housekeeping gene, are shown. Two technical replicates from two biological replicates are displayed. ***p-value < 0.001 compared to each gene in HET-1A cells.