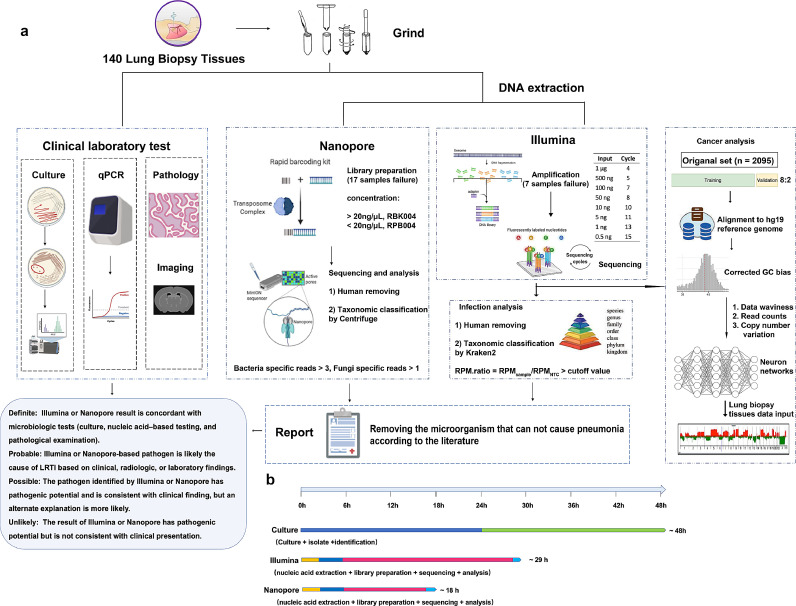
Fig. 1.
Schematic workflow of this study. (a) Lung biopsy samples analysis from 133 patients. Several microbiological tests were performed on these samples, including BALF culture, qPCR, ELISA, GeneXpert MTB/RIF, and histology. DNA was extracted and prepared for Illumina sequencing (seven samples showed library failure), Nanopore sequencing (17 samples showed library failure), and final analysis. The pathogen results of Illumina and Nanopore sequencing were adjudicated based on the literature from PubMed and their clinical condition. Three neural networks were pretrained by extracting features of human reads from 2,095 metagenomic next-generation sequencing results. The Illumina sequencing results were mapped to the human reference and then applied to the model. (b) The TAT of different methods. Abbreviations: BALF, bronchoalveolar lavage fluid; ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction; TAT, turnaround time.